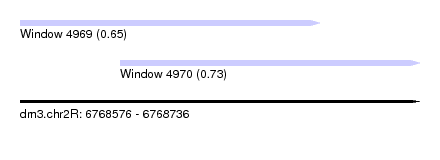
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,768,576 – 6,768,736 |
| Length | 160 |
| Max. P | 0.726649 |

| Location | 6,768,576 – 6,768,696 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.52156 |
| G+C content | 0.51154 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -14.94 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
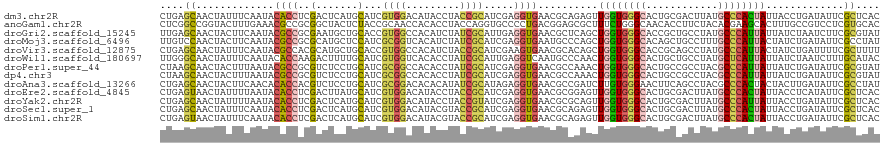
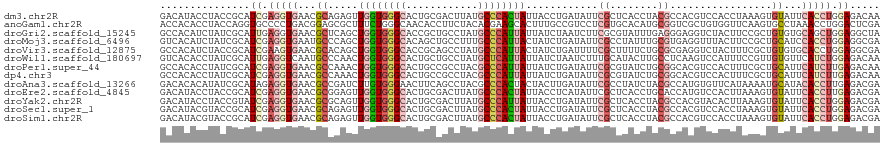
>dm3.chr2R 6768576 120 + 21146708 CUGAGCAACUAUUUCAAUACACCUCGACUCAUGCAUCGUGGACAUACCUACCGCAUCGAGGUGAACGCAGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUGAUAUUCGCUCAC .(((((.............((((((((....(((...((((......)))).)))))))))))....(((.(((((((((((..........)))))))))..)))))......))))). ( -42.10, z-score = -4.55, R) >anoGam1.chr2R 6813407 120 + 62725911 CUCGGCCGGUACUUUGAAACGCCGCGGCUACUCUACCGCAACCACACCUACCAGGUGCCCCUGACGGAGCGCUUUCUGGGCAACACCUUCUACACGAAGCACUUUGCCGUCCUCGUGCAC ..((((.(((.(((((.......((((........)))).......((((..((((((.((....)).))))))..))))..............))))).)))..))))........... ( -32.90, z-score = 0.05, R) >droGri2.scaffold_15245 4126870 120 + 18325388 UUGAGCAACUACUUCAAUACGCCGCGAAUGCUGCACCGUGGCCACAUCUAUCGCAUUGAGGUGAACGCUCAGCUGGUGGGCACCGCUGCCUAUGCCCAUUAUUAUCUAAUCUUCGCGUAU ((((((...(((((((((.(((((((..........))))))..........).)))))))))...)))))).(((((((((..........)))))))))................... ( -33.20, z-score = -0.03, R) >droMoj3.scaffold_6496 10979302 120 - 26866924 UUGUCCAACUACUUCAAUACGCCGCGCAUGCUCCAUCGCGGUCACAUCUAUCGCAUCGAGGUGAAUGCCCAGCUGGUGGGCACAGCUGCCUUUGCCCAUUACUAUCUGAUAUUCGCCUAU ....................((((((.(((...)))))))))................(((((((((..(((.(((((((((.((....)).)))))))))....))).))))))))).. ( -35.60, z-score = -1.40, R) >droVir3.scaffold_12875 11044611 120 + 20611582 CUGAGCAACUAUUUCAAUACGCCACGCAUGCUGCACCGUGGCCACAUCUACCGCAUCGAAGUGAACGCACAGCUGGUGGGCACCGCAGCCUAUGCCCAUUACUAUCUGAUUUUCGCUUUU ..((((...(((....))).((((((..........))))))..........)).))((((((((....(((.(((((((((..........)))))))))....)))...)))))))). ( -28.00, z-score = 0.79, R) >droWil1.scaffold_180697 528739 120 - 4168966 UUGGGCAACUAUUUCAAUACACCAAGACUUUUGCAUCGUGGUCACACCUAUCGCAUUGAGGUCAAUGCCCAACUGGUGGGCACUGCUGCCUAUGCUCAUUAUUAUCUAAUCUUUGCAUAC .((((((............(((((.((((..........)))).........((((((....)))))).....)))))((((....))))..))))))...................... ( -29.50, z-score = -1.02, R) >droPer1.super_44 366575 120 + 614636 CUAAGCAACUACUUUAAUACGCCGCGUCUCCUGCAUCGCGGCCACACCUAUCGCAUCGAGGUGAACGCCAAACUGGUGGGCACUGCCGCCUACGCCCAUUAUUAUCUGAUAUUCGCGUAU ....((..............((((((..........))))))..(((((.........)))))...))......(((((......)))))(((((....((((....))))...))))). ( -31.40, z-score = -0.36, R) >dp4.chr3 12183117 120 + 19779522 CUAAGCAACUACUUUAAUACGCCGCGUCUCCUGCAUCGCGGCCACACCUAUCGCAUCGAGGUGAACGCCAAACUGGUGGGCACUGCCGCCUACGCCCAUUAUUAUCUGAUAUUCGCGUAU ....((..............((((((..........))))))..(((((.........)))))...))......(((((......)))))(((((....((((....))))...))))). ( -31.40, z-score = -0.36, R) >droAna3.scaffold_13266 18265566 120 + 19884421 CUGAGCAACUACUUCAACACACCACGUCUCCUGCAUCGCGGACACACAUAUCGCAUAGAGGUGAACGCCGAUCUUGUGGGAACUUCAGCCUACGCCCACUACUACUUGAUAUUCGCCUAU ....((...................((.(((.((...))))).))..((((((..(((.(((....)))......(((((..............)))))..)))..))))))..)).... ( -21.54, z-score = 0.63, R) >droEre2.scaffold_4845 3582663 120 - 22589142 CUGAGUAACUAUUUUAAUACACCUCGACUUAUGCAUCGUGGACAUACCUACCGCAUCGAGGUGAACGCGGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUCAUAUUCGCUCAC .(((((.............((((((((....(((...((((......)))).)))))))))))...(.((...(((((((((..........)))))))))...)).)......))))). ( -39.60, z-score = -4.06, R) >droYak2.chr2R 18778724 120 - 21139217 CUGAGCAACUAUUUUAAUACACCUCGACUCAUGCAUCGUGGACAUACCUACCGUAUCGAGGUGAACGCGCAGUUGGUGGGCACUGCGACUUAUGCCCAUUAUUACCUGAUAUUCGCUCAC .(((((.............((((((((....(((...((((......)))).)))))))))))......(((.(((((((((..........)))))))))....)))......))))). ( -36.50, z-score = -2.92, R) >droSec1.super_1 4355052 120 + 14215200 CUGAGCAACUAUUUCAAUACACCUCGACUCAUGCAUCGUGGACAUACGUACCGCAUCGAGGUGAACGCAGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUGAUAUUCGCUCAC .(((((.............((((((((....(((..((((....))))....)))))))))))....(((.(((((((((((..........)))))))))..)))))......))))). ( -42.00, z-score = -4.06, R) >droSim1.chr2R 5358098 120 + 19596830 CUGAGUAACUAUUUCAAUACACCUCGACUCAUGCAUCGUGGACAUACGUACCGCAUCGAGGUGAACGCAGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUGAUAUUCGCUCAC .(((((.............((((((((....(((..((((....))))....)))))))))))....(((.(((((((((((..........)))))))))..)))))......))))). ( -39.60, z-score = -3.33, R) >consensus CUGAGCAACUAUUUCAAUACACCGCGACUCCUGCAUCGUGGACACACCUACCGCAUCGAGGUGAACGCCCAGCUGGUGGGCACUGCGGCCUAUGCCCAUUAUUAUCUGAUAUUCGCUUAC ....((.............((((..(.......)...((((.........)))).....))))..........((((((((............)))))))).............)).... (-14.94 = -14.70 + -0.24)
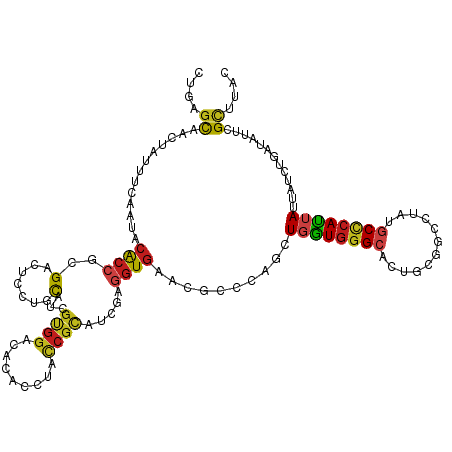
| Location | 6,768,616 – 6,768,736 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.12 |
| Shannon entropy | 0.58622 |
| G+C content | 0.52115 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
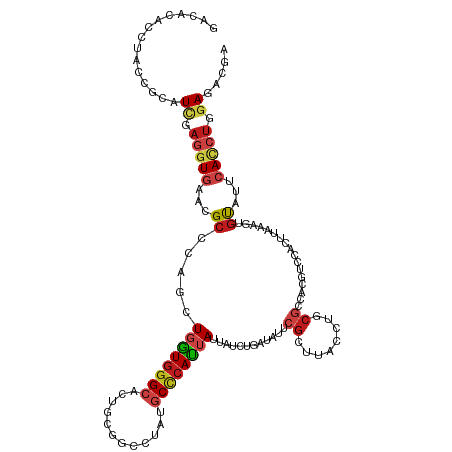
>dm3.chr2R 6768616 120 + 21146708 GACAUACCUACCGCAUCGAGGUGAACGCAGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUGAUAUUCGCUCACCUACGCCACGUCCACCUAAAGUGUAUUCACCUGGAGACAA ............(..((.(((((((..((.....(((((((...(((......((....((((....))))...)).......)))...))))))).....))..))))))).))..).. ( -33.42, z-score = -1.56, R) >anoGam1.chr2R 6813447 120 + 62725911 ACCACACCUACCAGGUGCCCCUGACGGAGCGCUUUCUGGGCAACACCUUCUACACGAAGCACUUUGCCGUCCUCGUGCACAUGCGGUCGCUGUGGUUCAAGUGCCUAAACCUGGACUCGA ..........((((((((.((....)).))......((((((.......((((((((.(((...(((((....)).)))..)))..))).)))))......)))))).))))))...... ( -39.02, z-score = -0.80, R) >droGri2.scaffold_15245 4126910 120 + 18325388 GCCACAUCUAUCGCAUUGAGGUGAACGCUCAGCUGGUGGGCACCGCUGCCUAUGCCCAUUAUUAUCUAAUCUUCGCGUAUUUGAGGGAGGUCUACUUCCGCUGUGUGCAGCUGGAGGCUA (((.((..((.(((.((((((....).))))).(((((((((..........))))))))).............)))))..)).(((((.....)))))((((....))))....))).. ( -38.30, z-score = 0.23, R) >droMoj3.scaffold_6496 10979342 120 - 26866924 GUCACAUCUAUCGCAUCGAGGUGAAUGCCCAGCUGGUGGGCACAGCUGCCUUUGCCCAUUACUAUCUGAUAUUCGCCUAUUUGCGUGAGGUUUACUUCCGCUGCAUCCACCUGGAGGCGA ..........((((.((.(((((.((((.(((.(((((((((.((....)).)))))))))....)))..............(((.((((....))))))).)))).))))).)).)))) ( -43.80, z-score = -2.27, R) >droVir3.scaffold_12875 11044651 120 + 20611582 GCCACAUCUACCGCAUCGAAGUGAACGCACAGCUGGUGGGCACCGCAGCCUAUGCCCAUUACUAUCUGAUUUUCGCUUUUCUGCGCGAGGUCUACUUUCGCUGUGUGCACCUGGAGGCGA ...........(((.((.(.(((..(((((((((((((((((..........)))))))))......(((((((((......))).)))))).......))))))))))).).)).))). ( -40.90, z-score = -1.15, R) >droWil1.scaffold_180697 528779 120 - 4168966 GUCACACCUAUCGCAUUGAGGUCAAUGCCCAACUGGUGGGCACUGCUGCCUAUGCUCAUUAUUAUCUAAUCUUUGCAUACUUGCCUCAAGUCCAUUUCCGUUGUGUUCAUCUGGAGACAA (((.........((((((....))))))(((...(((((((((.((....(((((..((((.....))))....)))))((((...)))).........)).)))))))))))).))).. ( -29.50, z-score = -0.83, R) >droPer1.super_44 366615 120 + 614636 GCCACACCUAUCGCAUCGAGGUGAACGCCAAACUGGUGGGCACUGCCGCCUACGCCCAUUAUUAUCUGAUAUUCGCGUAUCUGCGGCACGUCCACUUUCGCUGCAUUCAUCUUGAGACAA ............(..((((((((((.((......(((((((..((((((.(((((....((((....))))...)))))...)))))).)))))))......)).))))))))))..).. ( -43.60, z-score = -4.09, R) >dp4.chr3 12183157 120 + 19779522 GCCACACCUAUCGCAUCGAGGUGAACGCCAAACUGGUGGGCACUGCCGCCUACGCCCAUUAUUAUCUGAUAUUCGCGUAUCUGCGGCACGUCCACUUUCGCUGCAUUCAUCUUGAGACAA ............(..((((((((((.((......(((((((..((((((.(((((....((((....))))...)))))...)))))).)))))))......)).))))))))))..).. ( -43.60, z-score = -4.09, R) >droAna3.scaffold_13266 18265606 120 + 19884421 GACACACAUAUCGCAUAGAGGUGAACGCCGAUCUUGUGGGAACUUCAGCCUACGCCCACUACUACUUGAUAUUCGCCUAUCUACGCCAUGUGUUCAUAAAAUGCAUACACCUUGAGACGA ((.((((((...((.((((((((((...(((....(((((..............)))))......)))...))))))..)))).)).))))))))......................... ( -24.64, z-score = -0.42, R) >droEre2.scaffold_4845 3582703 120 - 22589142 GACAUACCUACCGCAUCGAGGUGAACGCGGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUCAUAUUCGCUCACCUGCACCAUGUCCACUUAAAGUGUAUUCACCUUGAGACGA ...........((..(((((((((((((.(((.(((((((((..........)))))))))....)))......((......))................))))..)))))))))..)). ( -38.00, z-score = -2.90, R) >droYak2.chr2R 18778764 120 - 21139217 GACAUACCUACCGUAUCGAGGUGAACGCGCAGUUGGUGGGCACUGCGACUUAUGCCCAUUAUUACCUGAUAUUCGCUCACCUACGCCACGUACACUUAAAGUGUAUUCACCUGGAGACGA ...........(((.((.((((((..(.((.((.(((((((...(((.....)))....((((....))))...))))))).)))))..((((((.....)))))))))))).)).))). ( -36.20, z-score = -2.12, R) >droSec1.super_1 4355092 120 + 14215200 GACAUACGUACCGCAUCGAGGUGAACGCAGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUGAUAUUCGCUCACCUACGCCACGUCCACCUAAAGUGUAUUCACCUGGAGACGA ...........((..((.(((((((..((.....(((((((...(((......((....((((....))))...)).......)))...))))))).....))..))))))).))..)). ( -35.22, z-score = -1.36, R) >droSim1.chr2R 5358138 120 + 19596830 GACAUACGUACCGCAUCGAGGUGAACGCAGAGUUGGUGGGCACUGCGACUUAUGCCCACUAUUACCUGAUAUUCGCUCACCUACGCCACGUCCACCUAAAGUGUAUUCACCUGGAGACGA ...........((..((.(((((((..((.....(((((((...(((......((....((((....))))...)).......)))...))))))).....))..))))))).))..)). ( -35.22, z-score = -1.36, R) >consensus GACACACCUACCGCAUCGAGGUGAACGCCCAGCUGGUGGGCACUGCGGCCUAUGCCCAUUAUUAUCUGAUAUUCGCUUACCUGCGCCACGUCCACUUAAAGUGUAUUCACCUGGAGACGA ...............((.(((((...((.....((((((((............))))))))............((........)).................))...))))).))..... (-13.82 = -14.38 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:41 2011