| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,745,545 – 6,745,639 |
| Length | 94 |
| Max. P | 0.659841 |

| Location | 6,745,545 – 6,745,639 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.70 |
| Shannon entropy | 0.51296 |
| G+C content | 0.49569 |
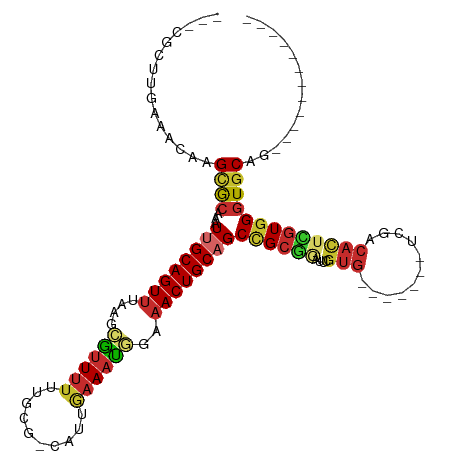
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
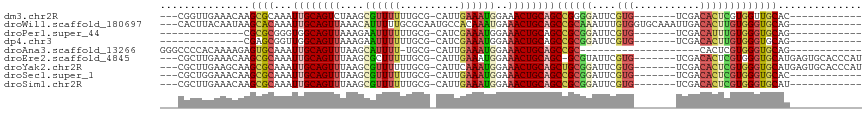
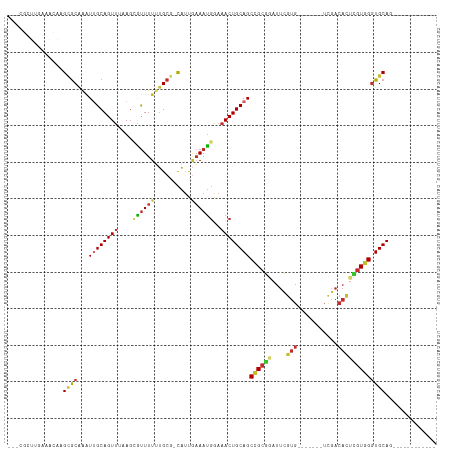
>dm3.chr2R 6745545 94 + 21146708 ---CGGUUGAAACAAGCGCAAAUUGCAGUCUAAGCGUUUUUUGCG-CAUUGAAAUGGAAACUGCAGCCGGGGAUUCGUG-------UCGACACUCGUGGUUGCAC------------ ---.........((((((((((.(((.......)))...))))))-).)))....(....)((((((((.((....(((-------....))))).)))))))).------------ ( -29.20, z-score = -0.75, R) >droWil1.scaffold_180697 495904 102 - 4168966 ---CACUUACAAUAAGCACAAAUUGCAGUUAAACAUUUUUGCGCAAUGCCACAAAUGAAACUGCAGCCGCAAAUUUGUGGUGCAAAUUGACACUUGUGGGUGCAG------------ ---(((((((((...(((.....))).(((((.(((((.((.((...)))).)))))....((((.(((((....)))))))))..)))))..)))))))))...------------ ( -29.00, z-score = -0.95, R) >droPer1.super_44 344746 83 + 614636 --------------CGCGCGGGUGGCAGUUAAAGAAUUUUUUGCG-CAUCGAAAUGGAAACUGCAGCCGCGGAUUCGUG-------UCGACAUUUGUGGGUGCAG------------ --------------.(((((((.................))))))-).............(((((.(((((((((((..-------.))).)))))))).)))))------------ ( -26.53, z-score = -0.58, R) >dp4.chr3 12160933 83 + 19779522 --------------CGAGCGGUUGGCAGUUAAAGAAUUUUUUGCG-CAUCGAAAUGGAAACUGCAGCCGCGGAUUCGUG-------UCGACACUUGUGGGUGCAG------------ --------------...(((((((.(((((.....(((((.....-....)))))...))))))))))))......(..-------((.((....)).))..)..------------ ( -23.80, z-score = -0.01, R) >droAna3.scaffold_13266 18241809 83 + 19884421 GGGCCCCACAAAAGAGUGCAAAUUGCAGUUUAAGCAUUUU-UGCG-CAUUGAAAUGGAAACUGCAGCCGC--------------------CACUCGUGGGUGCAG------------ ..(((((((....(((((....(((((((((...((((((-....-....)))))).)))))))))....--------------------)))))))))).))..------------ ( -31.20, z-score = -2.35, R) >droEre2.scaffold_4845 3559965 105 - 22589142 ---CGCUUGAAACAAGCGCAAAUUGCAGUUUAAGCGCUUUUUGCG-CAUUGAAAUGGAAACUGCAGC-GCGUAUUCGUG-------UCGACACUCGUGGGUGCAUGAGUGCACCCAU ---((((((...)))))).....((((((((..((((.....)))-)..........))))))))((-(((....))))-------)........(((((((((....))))))))) ( -39.40, z-score = -2.00, R) >droYak2.chr2R 18754687 106 - 21139217 ---CGCUUGAAGCAAGCGCAAAUUGCAGUUUAAGCGUUUUUUGCG-CAUUCAAAUGGAAACUGCAGCUGCGGAUUCGUG-------UCGACACUCGUGGGUGCAUGAGUGCACCCAU ---.(((((...)))))(((..(((((((((..((((.....)))-)..((....))))))))))).)))......(((-------....)))..(((((((((....))))))))) ( -39.20, z-score = -1.70, R) >droSec1.super_1 4331746 94 + 14215200 ---CGCUGGAAACAAGCGCAAAUUGCAGUUUAAGCGUUUUUUGCG-CAUUGAAAUGGAAACUGCAGCCGCGGAUUCGUG-------UCGACACUCGUGGGUGCAC------------ ---...((....)).(((((((..((.((....))))..))))))-)........(....)((((.((((((....(((-------....))))))))).)))).------------ ( -32.40, z-score = -1.19, R) >droSim1.chr2R 5339518 94 + 19596830 ---CGCUUGAAACAAGCGCAAAUUGCAGUUUAAGCGUUUUUUGCG-CAUUGAAAUGGAAACUGCAGCCGCGGAUUCGUG-------UCGACACUCGUGGGUGCAU------------ ---.........((((((((((..((.((....))))..))))))-).)))....(....)((((.((((((....(((-------....))))))))).)))).------------ ( -31.20, z-score = -0.90, R) >consensus ___CGCUUGAAACAAGCGCAAAUUGCAGUUUAAGCGUUUUUUGCG_CAUUGAAAUGGAAACUGCAGCCGCGGAUUCGUG_______UCGACACUCGUGGGUGCAG____________ ...............((((...((((((((....((((((..........))))))..))))))))((((((..(((..........)))...)))))))))).............. (-17.35 = -17.70 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:38 2011