| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,494,104 – 2,494,179 |
| Length | 75 |
| Max. P | 0.995483 |

| Location | 2,494,104 – 2,494,179 |
|---|---|
| Length | 75 |
| Sequences | 8 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Shannon entropy | 0.42093 |
| G+C content | 0.55363 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -17.17 |
| Energy contribution | -16.75 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
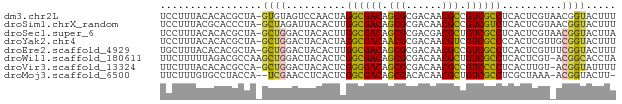
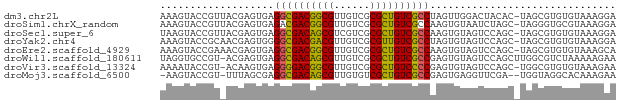
>dm3.chr2L 2494104 75 + 23011544 UCCUUUACACACGCUA-GUGUAGUCCAACUAGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUAACGGUACUUU .((.((((((((....-)))).........(((((((.(((......))).))))))).....)))).))...... ( -25.00, z-score = -2.88, R) >droSim1.chrX_random 149771 75 + 5698898 UCCUUUACGCACCCUA-GCUAGAUUACACUUGGCGACAGCGCGACAACGCCGUCGUCUCACUCGUAACGGUACUUU .((.((((((......-))............((((((.(((......))).))))))......)))).))...... ( -19.00, z-score = -1.37, R) >droSec1.super_6 4245616 75 - 4358794 UCCUUUACACACGCUA-GCUGGACUACACUUGGCGACAGCGCGACGACGCUGUCGCCUCACUCGUAACGGUACUUA .((.((((....(.((-(.....))))....((((((((((......))))))))))......)))).))...... ( -24.60, z-score = -2.56, R) >droYak2.chr4 88970 75 - 1374474 UCCUUUACACACGCUA-GCUGGACUACACUAGGCGACAACGCGACAACGUCGUCGCCCCACUCGUUGCGGUACUUU ........((.(((((-(.((.....)))))((((((.(((......))).)))))).........)))))..... ( -19.60, z-score = -1.02, R) >droEre2.scaffold_4929 1976492 75 - 26641161 UGCUUUACACACGCUA-GCUGGACUACACUUGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUUUCGGUACUUU ................-((((((........((((((.(((......))).))))))........))))))..... ( -21.09, z-score = -1.23, R) >droWil1.scaffold_180611 2732 75 + 8766 UUCUUUUUAGACGCCAAGCUGGACUACACUCGGCGACAGCGCGACAACGCUGUCGCCUCACUCGU-ACGGCACCUA ............(((.((.((.....)))).((((((((((......))))))))))........-..)))..... ( -26.30, z-score = -3.08, R) >droVir3.scaffold_13324 1039744 74 + 2960039 UUCUUUACACACGCCA-GCUGGACUACACUCGGGGACAGCGCGACAACGCCGUCCCCUCACUUGU-ACGGUAUUUU ................-((((.((.......((((((.(((......))).))))))......))-.))))..... ( -20.62, z-score = -1.90, R) >droMoj3.scaffold_6500 14041323 72 + 32352404 UUCUUUGUGCCUACCA--UCGAACCUCACUCGGCGACAGCGACACAACGCUGUCGCCUCGCUAAA-ACGGUACUU- ......(((((.....--..((.......))((((((((((......))))))))))........-..)))))..- ( -27.00, z-score = -5.29, R) >consensus UCCUUUACACACGCUA_GCUGGACUACACUCGGCGACAGCGCGACAACGCCGUCGCCUCACUCGU_ACGGUACUUU .................((((..........((((((.(((......))).))))))..........))))..... (-17.17 = -16.75 + -0.42)
| Location | 2,494,104 – 2,494,179 |
|---|---|
| Length | 75 |
| Sequences | 8 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Shannon entropy | 0.42093 |
| G+C content | 0.55363 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -21.28 |
| Energy contribution | -20.74 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
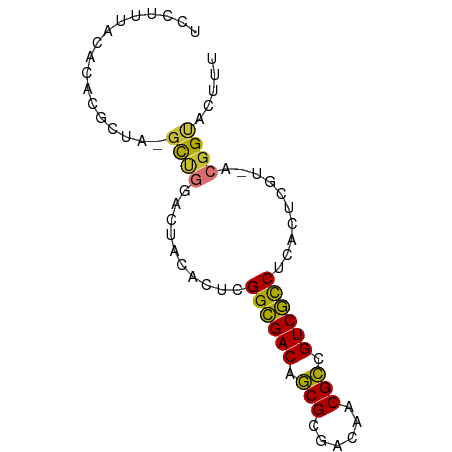
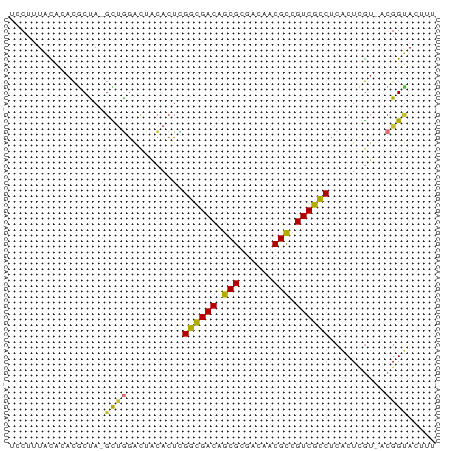
>dm3.chr2L 2494104 75 - 23011544 AAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCUAGUUGGACUACAC-UAGCGUGUGUAAAGGA ......(((((.(((.(.((((((((((((....))))))))))))).))))))(((((-......)))))..)). ( -32.40, z-score = -2.88, R) >droSim1.chrX_random 149771 75 - 5698898 AAAGUACCGUUACGAGUGAGACGACGGCGUUGUCGCGCUGUCGCCAAGUGUAAUCUAGC-UAGGGUGCGUAAAGGA ...(((((((((((..((.(.(((((((((....))))))))))))..))))))((...-.)))))))........ ( -27.70, z-score = -1.97, R) >droSec1.super_6 4245616 75 + 4358794 UAAGUACCGUUACGAGUGAGGCGACAGCGUCGUCGCGCUGUCGCCAAGUGUAGUCCAGC-UAGCGUGUGUAAAGGA ......((.(((((.((..(((((((((((....))))))))))).....(((.....)-))))...))))).)). ( -29.90, z-score = -2.85, R) >droYak2.chr4 88970 75 + 1374474 AAAGUACCGCAACGAGUGGGGCGACGACGUUGUCGCGUUGUCGCCUAGUGUAGUCCAGC-UAGCGUGUGUAAAGGA ....((((((........((((((((((((....))))))))))))....(((.....)-))))).)))....... ( -27.30, z-score = -1.50, R) >droEre2.scaffold_4929 1976492 75 + 26641161 AAAGUACCGAAACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGC-UAGCGUGUGUAAAGCA ....(((....(((.....(((((((((((....))))))))))).....(((.....)-)).)))..)))..... ( -25.10, z-score = -0.89, R) >droWil1.scaffold_180611 2732 75 - 8766 UAGGUGCCGU-ACGAGUGAGGCGACAGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUUGGCGUCUAAAAAGAA (((..(((..-........(((((((((((....)))))))))))((((........)))))))..)))....... ( -34.10, z-score = -3.69, R) >droVir3.scaffold_13324 1039744 74 - 2960039 AAAAUACCGU-ACAAGUGAGGGGACGGCGUUGUCGCGCUGUCCCCGAGUGUAGUCCAGC-UGGCGUGUGUAAAGAA ...(((((((-...(((..(((((((((((....)))))))))))((......))..))-).))).))))...... ( -29.10, z-score = -3.01, R) >droMoj3.scaffold_6500 14041323 72 - 32352404 -AAGUACCGU-UUUAGCGAGGCGACAGCGUUGUGUCGCUGUCGCCGAGUGAGGUUCGA--UGGUAGGCACAAAGAA -...((((((-(...((..((((((((((......))))))))))..)).......))--)))))........... ( -29.60, z-score = -3.29, R) >consensus AAAGUACCGU_ACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGC_UAGCGUGUGUAAAGGA ...................((((((((((......))))))))))............................... (-21.28 = -20.74 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:32 2011