
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,683,181 – 6,683,294 |
| Length | 113 |
| Max. P | 0.959384 |

| Location | 6,683,181 – 6,683,294 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Shannon entropy | 0.44351 |
| G+C content | 0.54514 |
| Mean single sequence MFE | -40.01 |
| Consensus MFE | -27.63 |
| Energy contribution | -27.18 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
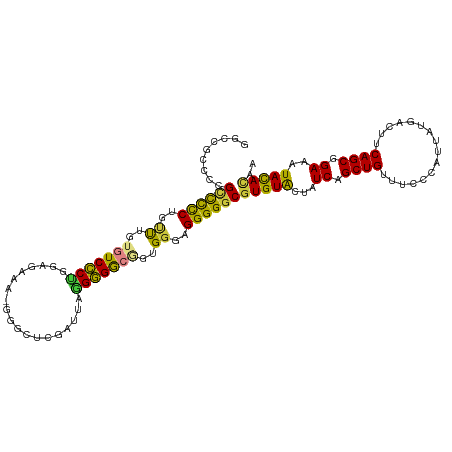
>dm3.chr2R 6683181 113 + 21146708 GGCCGCUCCGCCCUCUGUUUGUGUCCCUGGAGAAAAGGGCUCGAUUAGGGGACGAUGUGAGGGGGCGUGUACUAUCAGCUGUUUCCCAUUAUGACUACAGCGGAAAUACACAA (.((.(((((.((((((.(((.(.((((.......))))).))).)))))).))....))).)).)(((((...((.(((((.((.......))..))))).))..))))).. ( -39.20, z-score = -0.91, R) >droSim1.chr2R 5273681 113 + 19596830 GGCCGUUCCGCCCCCUGUUUGUGUCCCUGGAGAAAAGGGCUCGAUUUGGGGGCGAUGGGCGGGGGCUUGUACUAUCAGCUGUUUCCCAUUAUGACUUCAGCGGAAAUACACAA ..(((((..((((((...(((.(.((((.......))))).)))...))))))((((((..(.((((.(......))))).)..))))))........))))).......... ( -43.40, z-score = -1.07, R) >droSec1.super_1 4270538 112 + 14215200 GGCCGUUCCGCCCCCUGUUUGUGUCCCUGGAGAAAAGGGCUCGAUU-AGGGGCGAUGGGCGGGGGCGUGUACUAUCAGCUGUUUCCCAUUAUGACUUCAGCGGAAAUACACAA .....((((((((((((.(((.(.((((.......))))).))).)-))))).((((((..(.(((.((......))))).)..)))))).........))))))........ ( -42.00, z-score = -0.82, R) >droEre2.scaffold_4845 3496882 112 - 22589142 GGCCGCCCCGCCCCCUUUCUGUGUCCCUGGAGAAA-GGGCUCGAUUAGGGGGCGGUGGGAGGGGGCGUGUACUAUCAGCUGUUUCCCAUUAUGACUUCAGCGGAAAUACACAA ..((((.(((((((((...((.(.((((......)-)))).))...)))))))))(((((((.(((.((......))))).)))))))...........)))).......... ( -50.30, z-score = -2.07, R) >droYak2.chr2R 18691761 112 - 21139217 GGCUGCCCCGCCCCCUGUUUGUGUCCCCAGAGAAA-AGGCUCGAUUAGGGGGCAGUGGAAGGGGGCGUGUACUAUCAGCUGUUUUCCAUUAUGACUUCAGCGGAAAUACACAA ((.....))(((((((.....(((((((.(((...-...))).....))))))).....)))))))(((((...((.((((...((......))...)))).))..))))).. ( -43.00, z-score = -1.59, R) >dp4.chr3 3811565 98 - 19779522 ---AGCCCCGUUUCC-GCCUCUCUCUCUCUCGAAG-------AAUAAAGAGGUUGUGG--GGGGGCGUGUGUUAUCAGCUGUUGCCCAUUAUGACUUCAGCGGAAAUAUAU-- ---......((((((-((.(((.((......))))-------).....(((((..(((--..(((((...((.....))...))))).)))..))))).))))))))....-- ( -31.60, z-score = -1.40, R) >droPer1.super_4 794959 90 + 7162766 ---AGCCCCGUUCCC-GCCUCUCUCUC-------G-------AAGAAAGAGGUUGUGG--GGGG-CGUGUGUUAUCAGCUGUUGCCCAUUAUGACUUCAGCGGAAAUAUAU-- ---.(((((.(..(.-((((((.((..-------.-------..)).)))))).)..)--))))-)(((((((.((.((((................)))).)))))))))-- ( -30.59, z-score = -1.63, R) >consensus GGCCGCCCCGCCCCCUGUUUGUGUCCCUGGAGAAA_GGGCUCGAUUAGGGGGCGGUGGGAGGGGGCGUGUACUAUCAGCUGUUUCCCAUUAUGACUUCAGCGGAAAUACACAA .........((((((......(((((((.(((.......))).....)))))))......))))))(((((...((.((((................)))).))..))))).. (-27.63 = -27.18 + -0.45)


| Location | 6,683,181 – 6,683,294 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Shannon entropy | 0.44351 |
| G+C content | 0.54514 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6683181 113 - 21146708 UUGUGUAUUUCCGCUGUAGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCUCACAUCGUCCCCUAAUCGAGCCCUUUUCUCCAGGGACACAAACAGAGGGCGGAGCGGCC ..(((((((((.(((((..((......))..))))).)).)))))))((((((.....(((.........)))((((((((((....))).......))))))))).).))). ( -34.81, z-score = -1.22, R) >droSim1.chr2R 5273681 113 - 19596830 UUGUGUAUUUCCGCUGAAGUCAUAAUGGGAAACAGCUGAUAGUACAAGCCCCCGCCCAUCGCCCCCAAAUCGAGCCCUUUUCUCCAGGGACACAAACAGGGGGCGGAACGGCC ...((((((((.((((...((.......))..)))).)).)))))).......(((..((((((((........((((.......)))).........))))))))...))). ( -36.53, z-score = -1.01, R) >droSec1.super_1 4270538 112 - 14215200 UUGUGUAUUUCCGCUGAAGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCGCCCAUCGCCCCU-AAUCGAGCCCUUUUCUCCAGGGACACAAACAGGGGGCGGAACGGCC ..(((((((((.((((...((.......))..)))).)).)))))))......(((..((((((((-.......((((.......)))).........))))))))...))). ( -37.59, z-score = -1.26, R) >droEre2.scaffold_4845 3496882 112 + 22589142 UUGUGUAUUUCCGCUGAAGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCUCCCACCGCCCCCUAAUCGAGCCC-UUUCUCCAGGGACACAGAAAGGGGGCGGGGCGGCC ..(((((((((.((((...((.......))..)))).)).)))))))(((.((.....(((((((((..((...(((-(......)))).....)).))))))))))).))). ( -46.00, z-score = -2.41, R) >droYak2.chr2R 18691761 112 + 21139217 UUGUGUAUUUCCGCUGAAGUCAUAAUGGAAAACAGCUGAUAGUACACGCCCCCUUCCACUGCCCCCUAAUCGAGCCU-UUUCUCUGGGGACACAAACAGGGGGCGGGGCAGCC ..(((((((((.((((...((......))...)))).)).)))))))(((((........(((((((....(((...-...)))((....)).....)))))))))))).... ( -43.30, z-score = -2.64, R) >dp4.chr3 3811565 98 + 19779522 --AUAUAUUUCCGCUGAAGUCAUAAUGGGCAACAGCUGAUAACACACGCCCCC--CCACAACCUCUUUAUU-------CUUCGAGAGAGAGAGAGGC-GGAAACGGGGCU--- --.......((.((((..(((......)))..)))).))........(((((.--......(((((((.((-------(((...)))))))))))).-(....)))))).--- ( -32.90, z-score = -3.19, R) >droPer1.super_4 794959 90 - 7162766 --AUAUAUUUCCGCUGAAGUCAUAAUGGGCAACAGCUGAUAACACACG-CCCC--CCACAACCUCUUUCUU-------C-------GAGAGAGAGGC-GGGAACGGGGCU--- --.......((.((((..(((......)))..)))).))........(-((((--...(..((((((((..-------.-------..)))))))).-.)....))))).--- ( -33.60, z-score = -3.60, R) >consensus UUGUGUAUUUCCGCUGAAGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCUCCCACCGCCCCCUAAUCGAGCCC_UUUCUCCAGGGACACAAACAGGGGGCGGGGCGGCC ...((((((((.((((...((......))...)))).)).))))))(((((((.........(((.....................))).........)))))))........ (-20.73 = -21.57 + 0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:31 2011