| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,672,934 – 6,673,037 |
| Length | 103 |
| Max. P | 0.934521 |

| Location | 6,672,934 – 6,673,037 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 70.26 |
| Shannon entropy | 0.57616 |
| G+C content | 0.50335 |
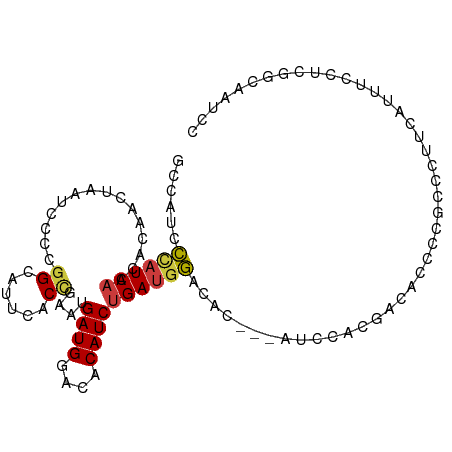
| Mean single sequence MFE | -16.87 |
| Consensus MFE | -9.96 |
| Energy contribution | -10.09 |
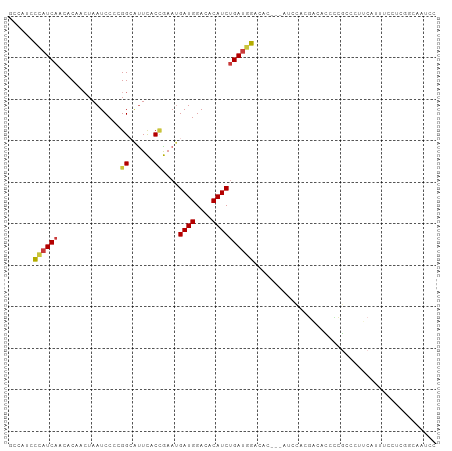
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
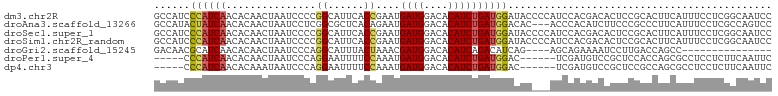
>dm3.chr2R 6672934 103 + 21146708 GCCAUCCCAUCAACACAACUAAUCCCCGGCAUUCACCGAAUGAUGGACACAUCUGAUGGAUACCCCAUCCACGACACUCCGCACUUCAUUUCCUCGGCAAUCC (((...((((((..............(((......)))...((((....))))))))))............((......))..............)))..... ( -19.80, z-score = -1.91, R) >droAna3.scaffold_13266 18164700 100 + 19884421 GCCAUACUAUCAACACAACUAAUCCUCGGCGCUCACAGAAUGAUGGACACAUCUGAUGGACAC---ACCCACAUCUUCCCGCCCUUCAUUUCCUCGCCAGUCC ...........................((((......((((((.((........((((.....---.....)))).......)).))))))...))))..... ( -15.26, z-score = -0.64, R) >droSec1.super_1 4252526 103 + 14215200 GCCAUCCCAUCAACACAACUAAUCCCCGGCAUUCACCGAAUGAUGGACACAUCUGAUGGAUACCCCAUCCACGACACUCCGCACUUCAUUUCCUCGGCAAUCC (((...((((((..............(((......)))...((((....))))))))))............((......))..............)))..... ( -19.80, z-score = -1.91, R) >droSim1.chr2R_random 1859708 103 + 2996586 GCCAUCCCAUCAACACAACUAAUCCCCGGCAUUCACCGAAUGAUGGACACAUCUGAUGGAUACCCCAUCCACGACACUCCGCACUUCAUUUCCUCGGCAAUCC (((...((((((..............(((......)))...((((....))))))))))............((......))..............)))..... ( -19.80, z-score = -1.91, R) >droGri2.scaffold_15245 5512859 84 + 18325388 GACAACGCAUCAACACAACUAAUCCCAGGCAUUUACUAAACGAUGGACACAUCAGACAUCAG----AGCAGAAAAUCCUUGACCAGCC--------------- ...........................(((...........((((....)))).....((((----(........))..)))...)))--------------- ( -8.00, z-score = 0.03, R) >droPer1.super_4 788054 92 + 7162766 -----CCCAUCAACACAACUAAUCCCAGGAAUUUUCCAAAUGAUGGACACAUCUGAUGGAC------UCGAUGUCCGCUCCACCAGCGCCUCCUCUUCAAUUC -----.((((((...............(((....)))....((((....))))))))))..------..((.(..((((.....))))...).))........ ( -17.70, z-score = -1.32, R) >dp4.chr3 3804671 92 - 19779522 -----CCCAUCAACACAAAUAAUCCCAGGAAUUUUCCAAAUGAUGGACACAUCUGAUGGAC------UCGAUGUCCGCUCCGCCAGCGCCUCCUCUUCAAUUC -----.((((((...............(((....)))....((((....))))))))))..------..((.(..((((.....))))...).))........ ( -17.70, z-score = -0.89, R) >consensus GCCAUCCCAUCAACACAACUAAUCCCCGGCAUUCACCGAAUGAUGGACACAUCUGAUGGACAC___AUCCACGACACCCCGCCCUUCAUUUCCUCGGCAAUCC ......((((((...............((......))....((((....))))))))))............................................ ( -9.96 = -10.09 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:28 2011