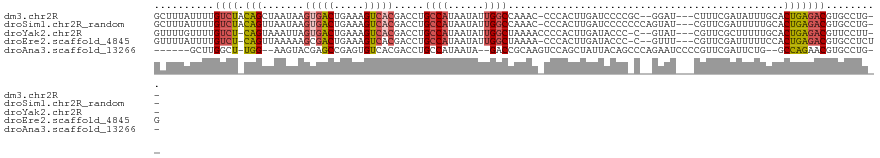
| Sequence ID | dm3.chr2R |
|---|---|
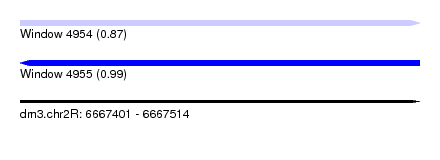
| Location | 6,667,401 – 6,667,514 |
| Length | 113 |
| Max. P | 0.986721 |

| Location | 6,667,401 – 6,667,514 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 74.21 |
| Shannon entropy | 0.45178 |
| G+C content | 0.46268 |
| Mean single sequence MFE | -31.61 |
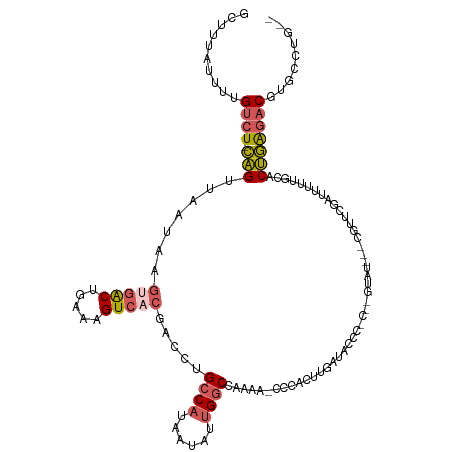
| Consensus MFE | -16.02 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
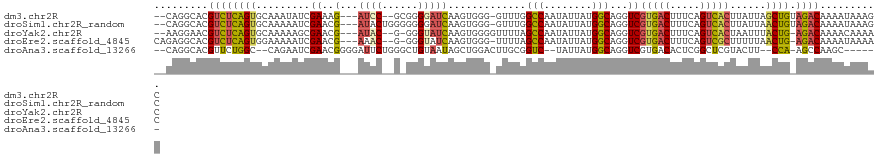
>dm3.chr2R 6667401 113 + 21146708 --CAGGCACGUCUCAGUGCAAAUAUCGAAAG---AUCC--GCGGGGAUCAAGUGGG-GUUUGGCCAAUAUUAUGGCAGGUCGUGACUUUCAGUCACUUAUUAGCUGUAGACAAAAUAAAGC --.......((((((((.(((((..((...(---((((--....)))))...))..-)))))((((......)))).....(((((.....)))))......)))).)))).......... ( -33.10, z-score = -2.42, R) >droSim1.chr2R_random 1843240 115 + 2996586 --CAGGCACGUCUCAGUGCAAAAAUCGAACG---AUACUGGGGGGGAUCAAGUGGG-GUUUGGCCAAUAUUAUGGCAGGUCGUGACUUUCAGUCACUUAUUAACUGUAGACAAAAUAAAGC --.......((((((((........(((((.---.((((.((.....)).))))..-)))))((((......)))).....(((((.....)))))......)))).)))).......... ( -28.80, z-score = -1.32, R) >droYak2.chr2R 18650127 112 - 21139217 --AAGGAACGUCUCAGUGCAAAAAGCGAACG---AUAC--G-GGGUAUCAAGUGGGGUUUUAGCCAAUAUUAUGGCAGGUCGUGACUUUCAGUCACUAAUUUACUG-AGACAAAACAAAAC --.......(((((((((........((.((---((((--.-..))))).............((((......)))).).))(((((.....))))).....)))))-)))).......... ( -30.10, z-score = -2.72, R) >droEre2.scaffold_4845 3478879 113 - 22589142 CAGAGGCACGUCUCAGUGGAAAAAUCGAACG---AAAC--G-GGGUAUCAAGUGGG-UUUUAGCCAAUAUUAUGGCAGGUCGUGACUUUCAGUCGCUUUUUAACUG-AGACAAAAUAAAAC .........((((((((.(((((...((.((---((((--.-.............)-)))).((((......)))).).))(((((.....)))))))))).))))-)))).......... ( -28.44, z-score = -1.79, R) >droAna3.scaffold_13266 18158698 106 + 19884421 --CAGGCACGUUCUGGC--CAGAAUCGAACGGGGAUUCUGGGCUGUAAUAGCUGGACUUGCGGUC--UAUUAUGGCAGGUCGUGACACUCGGCUCGUACUU--CCA-AGCCAAGC------ --..(.((((..((..(--(((((((.......))))))))(((((((((((((......))).)--)))))))))))..)))).)....((((.(.....--.).-))))....------ ( -37.60, z-score = -1.48, R) >consensus __CAGGCACGUCUCAGUGCAAAAAUCGAACG___AUAC__G_GGGUAUCAAGUGGG_UUUUGGCCAAUAUUAUGGCAGGUCGUGACUUUCAGUCACUUAUUAACUG_AGACAAAAUAAAAC .........((((((((.............................................((((......)))).....(((((.....)))))......)))).)))).......... (-16.02 = -16.06 + 0.04)
| Location | 6,667,401 – 6,667,514 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 74.21 |
| Shannon entropy | 0.45178 |
| G+C content | 0.46268 |
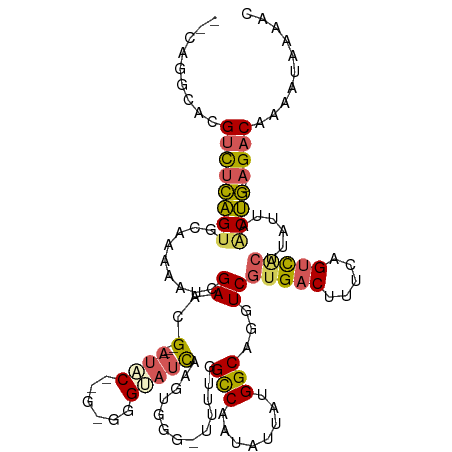
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6667401 113 - 21146708 GCUUUAUUUUGUCUACAGCUAAUAAGUGACUGAAAGUCACGACCUGCCAUAAUAUUGGCCAAAC-CCCACUUGAUCCCCGC--GGAU---CUUUCGAUAUUUGCACUGAGACGUGCCUG-- ..........((((...((.((((.(((((.....)))))((...((((......)))).....-.......(((((....--))))---)..))..)))).))....)))).......-- ( -25.60, z-score = -2.25, R) >droSim1.chr2R_random 1843240 115 - 2996586 GCUUUAUUUUGUCUACAGUUAAUAAGUGACUGAAAGUCACGACCUGCCAUAAUAUUGGCCAAAC-CCCACUUGAUCCCCCCCAGUAU---CGUUCGAUUUUUGCACUGAGACGUGCCUG-- ..........((((.((((......(((((.....))))).....((((......)))).....-................(((.((---(....)))..))).)))))))).......-- ( -21.10, z-score = -1.31, R) >droYak2.chr2R 18650127 112 + 21139217 GUUUUGUUUUGUCU-CAGUAAAUUAGUGACUGAAAGUCACGACCUGCCAUAAUAUUGGCUAAAACCCCACUUGAUACCC-C--GUAU---CGUUCGCUUUUUGCACUGAGACGUUCCUU-- ..........((((-((((......(((((.....)))))((.(.((((......)))).............(((((..-.--))))---)).))((.....)))))))))).......-- ( -30.30, z-score = -4.27, R) >droEre2.scaffold_4845 3478879 113 + 22589142 GUUUUAUUUUGUCU-CAGUUAAAAAGCGACUGAAAGUCACGACCUGCCAUAAUAUUGGCUAAAA-CCCACUUGAUACCC-C--GUUU---CGUUCGAUUUUUCCACUGAGACGUGCCUCUG ..........((((-((((..((((((((.(((((((....))..((((......)))).....-..............-.--.)))---)).))).)))))..))))))))......... ( -26.00, z-score = -2.98, R) >droAna3.scaffold_13266 18158698 106 - 19884421 ------GCUUGGCU-UGG--AAGUACGAGCCGAGUGUCACGACCUGCCAUAAUA--GACCGCAAGUCCAGCUAUUACAGCCCAGAAUCCCCGUUCGAUUCUG--GCCAGAACGUGCCUG-- ------((((((((-((.--.....))))))))))(.((((....((..(((((--(..(....).....))))))..))((((((((.......)))))))--)......)))).)..-- ( -36.70, z-score = -3.50, R) >consensus GCUUUAUUUUGUCU_CAGUUAAUAAGUGACUGAAAGUCACGACCUGCCAUAAUAUUGGCCAAAA_CCCACUUGAUACCC_C__GUAU___CGUUCGAUUUUUGCACUGAGACGUGCCUG__ ..........((((.(((.......(((((.....))))).....((((......))))..............................................)))))))......... (-12.96 = -13.60 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:28 2011