
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,664,243 – 6,664,392 |
| Length | 149 |
| Max. P | 0.511164 |

| Location | 6,664,243 – 6,664,392 |
|---|---|
| Length | 149 |
| Sequences | 5 |
| Columns | 177 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Shannon entropy | 0.36277 |
| G+C content | 0.38659 |
| Mean single sequence MFE | -41.51 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
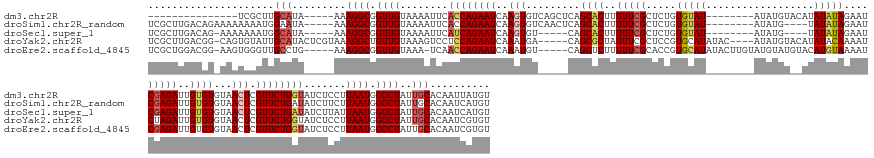
>dm3.chr2R 6664243 149 - 21146708 ---------------UCGCUUGCAUA-----AAAGGCGGUUGUAAAAUUCACCAGAAUCAAGUGUCAGCUCAGCACUUUUUCGCUCUGUGUAU--------AUAUGUACAUAUAUAGAAUCGAGAUUGUGUGUAACUCGUUCUGGUAUCUCCUUAAUGGCCUAUUGCACAAUUAUGU ---------------.....((((..-----..((((.((((........((((((((..(((....((...((((..(((((.(((((((((--------........)))))))))..)))))..)))))).))).)))))))).......)))).))))..))))......... ( -40.06, z-score = -2.47, R) >droSim1.chr2R_random 1839671 160 - 2996586 UCGCUUGACAGAAAAAAAAUGGAAUA-----AAAGGCGGUUGUAAAAUUCACCAGAAUCAAGUGUCAACUCAGCACUUUUUCGCUCUGUGUAU--------AUAUG----UAUAUAGAAUCGAGAUUGUGUGUAACUCGUUCUGAUAUCUUCUUAAUGGCCUAUUGCACAAUCAUGU ..((((....................-----..))))((((((..(((...(((.....((((((((((...((((..(((((.(((((((((--------....)----))))))))..)))))..)))).......))..))))).))).....)))...)))..)))))).... ( -33.05, z-score = -0.31, R) >droSec1.super_1 4243846 154 - 14215200 UCGCUUGACAG-AAAAAAAUGGCAUA-----AAAGGCGGUUGUAAAAUUCAUCAGAAUCAAGUGU-----CAGCACUUUUUCGCUCUGUGUAU--------AUAUG----UAUAUAGAAUCGAGAUUGUGUGUAACUCGUUCUGAUAUCUUAUUAAUGGCCUAUUGCACAAUCAUGU ..((.(((((.-.......))(((..-----..((((.((((........((((((((..(((..-----..((((..(((((.(((((((((--------....)----))))))))..)))))..))))...))).)))))))).......)))).))))..)))....))).)) ( -41.66, z-score = -2.82, R) >droYak2.chr2R 18647081 167 + 21139217 UCGCUUGACGG-CAGUGUAUUGCAUACUCGUAAAGGCUGUUGUAAAGUCCUCCAGAAUCAAAUGA-----CAGCGCUAUUUCGCUCCGUGCAUAUAC----AUAUGUACAUAUACAAAAUCUAGAUUGUGUGUAACUCGUUCUGGUAUCUCCUUAAUGGCCUAUUGCACAAUCGUGU .......((((-..(((((((((......))))(((((((((...((....(((((((.......-----.((((......))))..(((((((...----.)))))))((((((((........)))))))).....)))))))...))...)))))))))..)))))..)))).. ( -44.10, z-score = -1.70, R) >droEre2.scaffold_4845 3475776 165 + 22589142 UCGCUGGACGG-AAGUGGGUUGCCUG-----AAAGGCGGUUGUAAA-UCAACCAGAAUCAAAUGU-----CAGCUCUUUUUCGCACCGUGCAUAUACUUGUAUGUAUGUACAUGUAAAAUCGAGAUUGUGUGUAACUCGUUCUGGUAUCUCCUUAAUGGCCUAUUGCACAAUCGUGU ..(((((((..-..)).(((((((..-----...)))(((((....-.)))))..)))).....)-----))))........((((.(((((((((......)))))))))............(((((((((((..(((((..((.....))..)))))..)).))))))))))))) ( -48.70, z-score = -1.95, R) >consensus UCGCUUGACAG_AAAUAAAUUGCAUA_____AAAGGCGGUUGUAAAAUUCACCAGAAUCAAGUGU_____CAGCACUUUUUCGCUCUGUGUAU________AUAUGU__AUAUAUAGAAUCGAGAUUGUGUGUAACUCGUUCUGGUAUCUCCUUAAUGGCCUAUUGCACAAUCAUGU .................................((((.((((........((((((((..............((((...........))))...................((((((.(((....))).))))))....)))))))).......)))).))))............... (-22.16 = -22.04 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:25 2011