
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,660,001 – 6,660,122 |
| Length | 121 |
| Max. P | 0.728363 |

| Location | 6,660,001 – 6,660,122 |
|---|---|
| Length | 121 |
| Sequences | 6 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 72.70 |
| Shannon entropy | 0.49420 |
| G+C content | 0.50504 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -14.63 |
| Energy contribution | -17.13 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
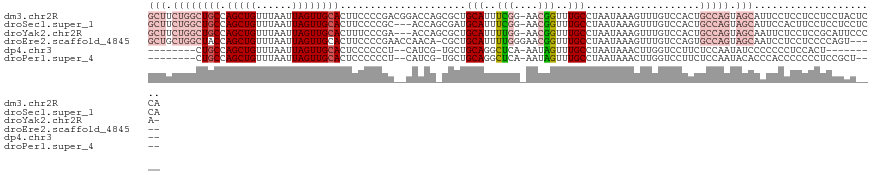
>dm3.chr2R 6660001 121 + 21146708 GCUUCUGGCUGCCAGCUGUUUAAUUAGUUGCACUUCCCCGACGGACCAGCGCUGCAUUUCGG-AACGGUUUGCCUAAUAAAGUUUGUCCACUGCCAGUAGCAUUCCUCCUCCUCCUACUCCA (((.((((((((.(((((......))))))))..........((((.(((...(((..(((.-..)))..)))........))).))))...))))).)))..................... ( -30.40, z-score = -1.02, R) >droSec1.super_1 4239676 118 + 14215200 GCUUCUGGCUGCCAGCUGUUUAAUUAGUUGCACUUCCCCGC---ACCAGCGAUGCAUUUCGG-AACGGUUUGCCUAAUAAAGUUUGUCCACUGCCAGUAGCAUUCCACUUCCUCCUCCUCCA (((.(((((.((.((((((((.......((((......(((---....))).)))).....)-))))))).)).......(((......)))))))).)))..................... ( -27.80, z-score = -1.21, R) >droYak2.chr2R 18642613 117 - 21139217 GCUUCUGGCUGCCAGCUGUUUAAUUAGUUGCACUUUCCCGA---ACCAGCGCUGCAUUUUGG-AACGGUUUGCCUAAUAAAGUUUGUCCACUGCCAGUAGCAAUUCUCCUCCGCAUUCCCA- (((.(((((.((.((((((((.....((.((.((.......---...)).)).))......)-))))))).)).......(((......)))))))).)))....................- ( -25.40, z-score = 0.19, R) >droEre2.scaffold_4845 3471818 116 - 22589142 GCUGCUGGCUACCAGCUGUUUAAUUAGUUGCACUUCCCCGAACCAACA-CGCUGCAUUUUGGGAACGGUUUGCCUAAUAAAGUUUGUCCAGUGCCAGUAGCAAUCCUCCUCCCCAGU----- (((((((((...((((((......)))))).(((....(((((.....-....(((..(((....)))..)))........)))))...))))))))))))................----- ( -31.63, z-score = -1.35, R) >dp4.chr3 3794370 101 - 19779522 --------CUGCCAGCUGUUUAAUUAGUUGCACUCCCCCCU--CAUCG-UGCUGCAGGCUCA-AAUAGUUUGCCUAAUAAACUUGGUCCUUCUCCAAUAUCCCCCCCUCCACU--------- --------..(((((..(((((.((((..((((........--....)-))).(((((((..-...)))))))))))))))))))))..........................--------- ( -18.20, z-score = -2.14, R) >droPer1.super_4 777777 106 + 7162766 --------CUGCCAGCUGUUUAAUUAGUUGCACUCCCCCCU--CAUCG-UGCUGCAGGCUCA-AAUAGUUUGCCUAAUAAACUUGGUCCUUCUCCAAUACACCCACCCCCCCUCCGCU---- --------.....(((.(((((.((((..((((........--....)-))).(((((((..-...))))))))))))))))((((.......))))..................)))---- ( -18.70, z-score = -1.76, R) >consensus GCUUCUGGCUGCCAGCUGUUUAAUUAGUUGCACUUCCCCGA__CACCA_CGCUGCAUUUUGG_AACGGUUUGCCUAAUAAAGUUUGUCCACUGCCAGUAGCAAUCCUCCUCCUCCUCC____ (((.((((((((.(((((......)))))))).....................(((..(((....)))..)))...................))))).)))..................... (-14.63 = -17.13 + 2.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:24 2011