| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,648,066 – 6,648,240 |
| Length | 174 |
| Max. P | 0.997920 |

| Location | 6,648,066 – 6,648,240 |
|---|---|
| Length | 174 |
| Sequences | 3 |
| Columns | 188 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Shannon entropy | 0.16607 |
| G+C content | 0.44465 |
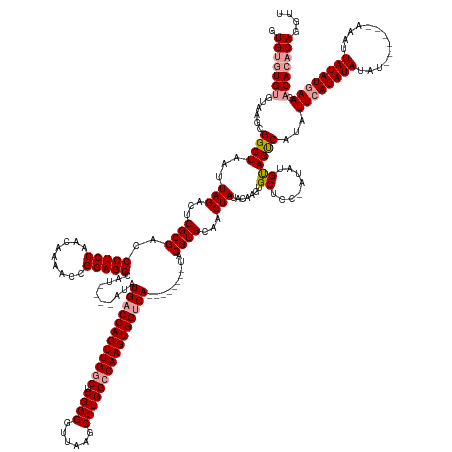
| Mean single sequence MFE | -61.61 |
| Consensus MFE | -49.56 |
| Energy contribution | -51.12 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
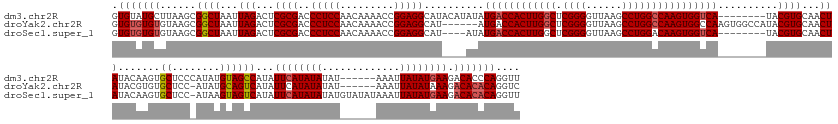
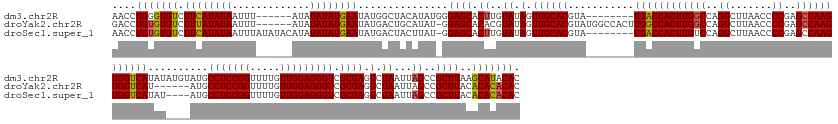
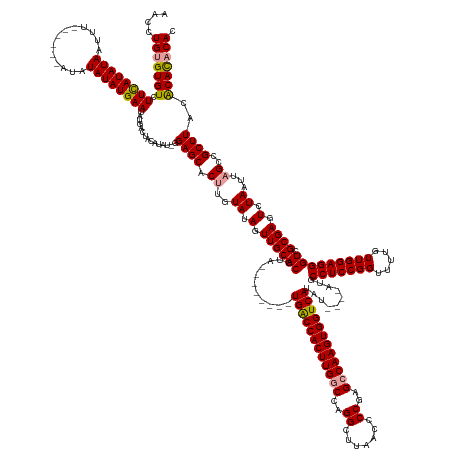
>dm3.chr2R 6648066 174 + 21146708 GUGUAUGCUUAAGCGGCUAAUUAGACUCGCGACCCUCCAACAAAACCGGAGGCAUACAUAUAUGACCACUUGGCUCGGGGUUAAGCCUGGCCAAGUGGUCA--------UACGUGCAACUAUACAAGUGCUCCCAUAUGUAGCCAUAUUCAUAUAUAU------AAAUUAUAUGAAGACACCCAGGUU ((((..((....))(((....(((...((((..(((((.........)))))........((((((((((((((.((((......))))))))))))))))--------))))))...)))((((.(((....))).)))))))...((((((((...------....)))))))).))))....... ( -61.70, z-score = -4.43, R) >droYak2.chr2R 18629045 175 - 21139217 GUGUGUGUGUAAGCGGCUAAUUAGACUCGCGACCCUCCAACAAAACCGGAGGCAU------AUGACCACUUGGCUCGGGGUUAAGCCUGGCCAAGUGGCCAAGUGGCCAUACGUGCAACUAUACGUGUGCUCC-AUAUGCAGUCAUAUUCAUAUAUAU------AAAUUAUAUAAAGACACACAGGUC .(((((((.(..((((((....)).).)))(((.((((.........))))((((------(((.(((((((((.((((......)))))))))))))......(((.(((((((......)))))))))).)-)))))).))).......((((((.------....)))))).).))))))).... ( -63.50, z-score = -3.53, R) >droSec1.super_1 4228023 175 + 14215200 GUGUGUGUGUAAGCGGCUAAUUAGACUCGCGACCCUCCAACAAAACCGGAGGCAU----AUAUGACCACUUGGCUCGGGGUUAAGCCUGGACAAGUGGUCA--------UACGUGCAACUAUACAAGUGCUCC-AUAAGUAGUCAUAUUCAUAUAUAUGUAUAUAAAUUAUAUGAAGACACACAGGUU .(.(((((((....(((((.(((....(((....((((.........))))(((.----.((((((((((((..(((((......))))).))))))))))--------))..)))..........)))....-.))).)))))...((((((((.............)))))))).))))))).).. ( -59.62, z-score = -3.82, R) >consensus GUGUGUGUGUAAGCGGCUAAUUAGACUCGCGACCCUCCAACAAAACCGGAGGCAU____AUAUGACCACUUGGCUCGGGGUUAAGCCUGGCCAAGUGGUCA________UACGUGCAACUAUACAAGUGCUCC_AUAUGUAGUCAUAUUCAUAUAUAU______AAAUUAUAUGAAGACACACAGGUU .(((((((......((((...(((...((((..(((((.........)))))..........((((((((((((.((((......))))))))))))))))..........))))...))).......((........))))))...((((((((.............)))))))).))))))).... (-49.56 = -51.12 + 1.56)
| Location | 6,648,066 – 6,648,240 |
|---|---|
| Length | 174 |
| Sequences | 3 |
| Columns | 188 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Shannon entropy | 0.16607 |
| G+C content | 0.44465 |
| Mean single sequence MFE | -61.17 |
| Consensus MFE | -49.40 |
| Energy contribution | -49.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6648066 174 - 21146708 AACCUGGGUGUCUUCAUAUAAUUU------AUAUAUAUGAAUAUGGCUACAUAUGGGAGCACUUGUAUAGUUGCACGUA--------UGACCACUUGGCCAGGCUUAACCCCGAGCCAAGUGGUCAUAUAUGUAUGCCUCCGGUUUUGUUGGAGGGUCGCGAGUCUAAUUAGCCGCUUAAGCAUACAC .......((((.((((((((....------...))))))))...(((((...((...((.((((((.....((((..((--------((((((((((((..((.......))..))))))))))))))..))))..(((((((.....)))))))...)))))))).)))))))((....))..)))) ( -62.00, z-score = -3.09, R) >droYak2.chr2R 18629045 175 + 21139217 GACCUGUGUGUCUUUAUAUAAUUU------AUAUAUAUGAAUAUGACUGCAUAU-GGAGCACACGUAUAGUUGCACGUAUGGCCACUUGGCCACUUGGCCAGGCUUAACCCCGAGCCAAGUGGUCAU------AUGCCUCCGGUUUUGUUGGAGGGUCGCGAGUCUAAUUAGCCGCUUACACACACAC .....(((((.((((((((.(.((------((((......)))))).)..))))-)))))))))(((.(((.((.(((((((((((((((((....)))).(((((......)))))))))))))))------)))(((((((.....)))))))(.(....).)......)).))))))........ ( -65.60, z-score = -3.96, R) >droSec1.super_1 4228023 175 - 14215200 AACCUGUGUGUCUUCAUAUAAUUUAUAUACAUAUAUAUGAAUAUGACUACUUAU-GGAGCACUUGUAUAGUUGCACGUA--------UGACCACUUGUCCAGGCUUAACCCCGAGCCAAGUGGUCAUAU----AUGCCUCCGGUUUUGUUGGAGGGUCGCGAGUCUAAUUAGCCGCUUACACACACAC ....((((((((((((((....((((((.(((....))).)))))).....)))-)))).....(((.(((.((..(((--------(((((((((.....(((((......)))))))))))))))))----...(((((((.....)))))))(.(....).)......)).))))))))))))). ( -55.90, z-score = -3.25, R) >consensus AACCUGUGUGUCUUCAUAUAAUUU______AUAUAUAUGAAUAUGACUACAUAU_GGAGCACUUGUAUAGUUGCACGUA________UGACCACUUGGCCAGGCUUAACCCCGAGCCAAGUGGUCAUAU____AUGCCUCCGGUUUUGUUGGAGGGUCGCGAGUCUAAUUAGCCGCUUACACACACAC ....(((((((.((((((((.............))))))))...............((((.((..((.(.((((((...........((((((((((((..((.......))..))))))))))))..........(((((((.....))))))))).)))).).))...))..))))..))))))). (-49.40 = -49.52 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:21 2011