| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,636,171 – 6,636,267 |
| Length | 96 |
| Max. P | 0.947737 |

| Location | 6,636,171 – 6,636,267 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.34823 |
| G+C content | 0.60271 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -27.40 |
| Energy contribution | -28.65 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
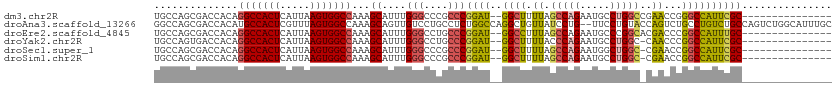
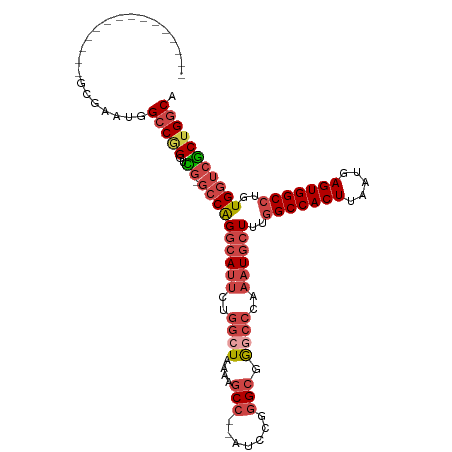
>dm3.chr2R 6636171 96 + 21146708 UGCCAGCGACCACAGGCCACUCAUUAAGUGGCCAAAGCAUUUGGGCCCGCCCGGAU--GGCUUUUAGCCAGAAUGCCUGGCCGAACCGGGCCAUUCGC--------------- .....((((.....(((((((.....)))))))..........((((((((.....--))).((..(((((.....)))))..))..)))))..))))--------------- ( -41.50, z-score = -2.28, R) >droAna3.scaffold_13266 18126966 111 + 19884421 GGCCAGCGACCACAUGCCACUCGUUUAGUGGCCAAAGCAGUUGUCCUGCCUCUGGCCAGGCUGUUAUCCUG--UUCCUGUACCAGUCUGCCUGUCUGCCAGUCUGGCAUUUGC ((((((.(.......((((((.....))))))....((((.....))))).))))))((((((.(((....--.....))).)))))).......((((.....))))..... ( -36.20, z-score = -0.65, R) >droEre2.scaffold_4845 3447478 96 - 22589142 UGCCAGCGACCACAGGCCACUCAUUAAGUGGCCAAAGCAUUUGGGCCUGCCCGGAU--GGCCUUUAGCCAGAAUGCCCGGCACGACCCGGCCAUUUGC--------------- .(((.(((....).(((((((.....)))))))...))....((((.((((.((.(--(((.....)))).....)).)))).).)))))).......--------------- ( -38.10, z-score = -1.73, R) >droYak2.chr2R 18617030 95 - 21139217 UGCCAGUGACCACAGGCCACUCAUUAAGUGGCCAAAGCAUUUGGGCCUGCCCGGAU--GGCUUUUACCCAGAAUGCCUGGC-CAACCCGGCCAUUCGC--------------- .....((((.....(((((((.....)))))))...(((((((((...(((.....--))).....))))).)))).((((-(.....))))).))))--------------- ( -36.50, z-score = -1.64, R) >droSec1.super_1 4216502 95 + 14215200 UGCCAGCGACCACAGGCCACUCAUUAAGUGGCCAAAGCAUUUGGGCCCGCCCGGAU--GGCUUUUAGCCAGAAUGGCUGGC-CGAACCGGCCAUUCGC--------------- .....((.......(((((((.....)))))))((((((((((((....)))))))--.)))))..))..((((((((((.-....))))))))))..--------------- ( -45.20, z-score = -3.49, R) >droSim1.chr2R 5242988 95 + 19596830 UGCCAGCGACCACAGGCCACUCAUUAAGUGGCCAAAGCAUUUGGGCCCGCCCGGAU--GGCUUUUAGCCAGAAUGCCUGGC-CGAACCGGCCAUUCGC--------------- .....((((.....(((((((.....)))))))...(((((((((....)))...(--(((.....)))))))))).((((-((...)))))).))))--------------- ( -41.80, z-score = -2.91, R) >consensus UGCCAGCGACCACAGGCCACUCAUUAAGUGGCCAAAGCAUUUGGGCCCGCCCGGAU__GGCUUUUAGCCAGAAUGCCUGGC_CAACCCGGCCAUUCGC_______________ .(((.((.......(((((((.....)))))))...))(((((((....)))))))..))).........(((((.((((......)))).)))))................. (-27.40 = -28.65 + 1.25)
| Location | 6,636,171 – 6,636,267 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.34823 |
| G+C content | 0.60271 |
| Mean single sequence MFE | -45.65 |
| Consensus MFE | -28.46 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
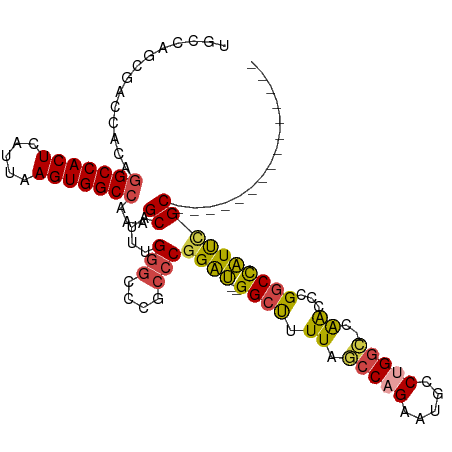
>dm3.chr2R 6636171 96 - 21146708 ---------------GCGAAUGGCCCGGUUCGGCCAGGCAUUCUGGCUAAAAGCC--AUCCGGGCGGGCCCAAAUGCUUUGGCCACUUAAUGAGUGGCCUGUGGUCGCUGGCA ---------------((((..(((((.....((((((.....))))))....(((--.....)))))))).....((...((((((((...)))))))).))..))))..... ( -43.40, z-score = -1.19, R) >droAna3.scaffold_13266 18126966 111 - 19884421 GCAAAUGCCAGACUGGCAGACAGGCAGACUGGUACAGGAA--CAGGAUAACAGCCUGGCCAGAGGCAGGACAACUGCUUUGGCCACUAAACGAGUGGCAUGUGGUCGCUGGCC ((...((((((.(((.(.....).))).))))))......--((((((.((((((.((((((((.(((.....)))))))))))(((.....)))))).))).))).))))). ( -45.70, z-score = -2.81, R) >droEre2.scaffold_4845 3447478 96 + 22589142 ---------------GCAAAUGGCCGGGUCGUGCCGGGCAUUCUGGCUAAAGGCC--AUCCGGGCAGGCCCAAAUGCUUUGGCCACUUAAUGAGUGGCCUGUGGUCGCUGGCA ---------------((...((((((((((.((((.((.....((((.....)))--).)).)))))))))....((...((((((((...)))))))).)))))))...)). ( -47.10, z-score = -2.12, R) >droYak2.chr2R 18617030 95 + 21139217 ---------------GCGAAUGGCCGGGUUG-GCCAGGCAUUCUGGGUAAAAGCC--AUCCGGGCAGGCCCAAAUGCUUUGGCCACUUAAUGAGUGGCCUGUGGUCACUGGCA ---------------.......(((((..((-((((((((((.(((((....(((--.....)))..)))))))))))..((((((((...))))))))..))))))))))). ( -46.40, z-score = -2.54, R) >droSec1.super_1 4216502 95 - 14215200 ---------------GCGAAUGGCCGGUUCG-GCCAGCCAUUCUGGCUAAAAGCC--AUCCGGGCGGGCCCAAAUGCUUUGGCCACUUAAUGAGUGGCCUGUGGUCGCUGGCA ---------------.......((((((..(-(((.(((....((((.....)))--)....))).)))).....(((..((((((((...))))))))...))).)))))). ( -44.40, z-score = -1.77, R) >droSim1.chr2R 5242988 95 - 19596830 ---------------GCGAAUGGCCGGUUCG-GCCAGGCAUUCUGGCUAAAAGCC--AUCCGGGCGGGCCCAAAUGCUUUGGCCACUUAAUGAGUGGCCUGUGGUCGCUGGCA ---------------.......(((((..((-((((((((((..((((....(((--.....))).))))..))))))..((((((((...))))))))..))))))))))). ( -46.90, z-score = -2.46, R) >consensus _______________GCGAAUGGCCGGGUCG_GCCAGGCAUUCUGGCUAAAAGCC__AUCCGGGCGGGCCCAAAUGCUUUGGCCACUUAAUGAGUGGCCUGUGGUCGCUGGCA ...............((.(.((((((......(((((.....)))))..(((((.......(((....)))....)))))(((((((.....)))))))..)))))).).)). (-28.46 = -29.13 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:16 2011