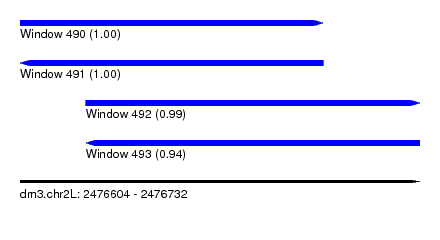
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,476,604 – 2,476,732 |
| Length | 128 |
| Max. P | 0.999534 |

| Location | 2,476,604 – 2,476,701 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 84.33 |
| Shannon entropy | 0.29492 |
| G+C content | 0.42156 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -4.49 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
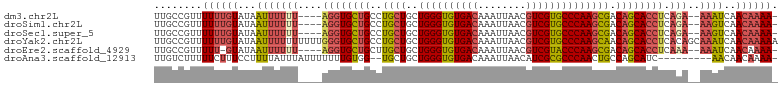
>dm3.chr2L 2476604 97 + 23011544 UUGCCGUUUUUUGUAUAAUUUUUU----AGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAAUCAACAAAA- ........((((((...(((((((----((((((((..((((..((((..((((........))))..)))))))).)))))))).)))--))))..))))))- ( -38.20, z-score = -5.13, R) >droSim1.chr2L 2438607 97 + 22036055 UUGCCGUUUUUUGUAUAAUUUUUU----AGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAGUCAACAAAA- ........((((((...(((((((----((((((((..((((..((((..((((........))))..)))))))).)))))))).)))--))))..))))))- ( -38.20, z-score = -4.77, R) >droSec1.super_5 650230 97 + 5866729 UUGCCGUUUUUUGUAUAAUUUUUU----AGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAGUCAACAAAA- ........((((((...(((((((----((((((((..((((..((((..((((........))))..)))))))).)))))))).)))--))))..))))))- ( -38.20, z-score = -4.77, R) >droYak2.chr2L 2467727 104 + 22324452 UUGCCGUUUUUUGUAUAAUUUUUUUUUUGGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCAACAGCACCUCACAGCAAAUCAACAAAAA .......(((((((...((((.......((((((((..((((..((((..((((........))))..)))))))).))))))))......))))..))))))) ( -34.82, z-score = -2.91, R) >droEre2.scaffold_4929 2521044 96 + 26641161 UUGCCGUUUUU-GUAUAAUUUUUU----AGGUGCUGCUUGCUGCUGGGUGUGACAAAUUAACGUCGUACCCAAGCGACAGCACCUCAAA--AAAUCAACAAAA- .......((((-((...(((((((----((((((((.(((((..((((((((((........))))))))))))))))))))))).)))--))))..))))))- ( -38.70, z-score = -6.02, R) >droAna3.scaffold_12913 218966 92 - 441482 UUGUCUUUUUCUUUCCUUUUAUUUAUUUUUUUGUGG--UGCUGCUGGGUGUGACAAAUUAACAUCGCGCCCAACUGCCAGCAUC---------AACAACAAAA- .........................((((.((((((--(((((((((((((((..........)))))))))...).)))))))---------.)))).))))- ( -25.30, z-score = -3.36, R) >consensus UUGCCGUUUUUUGUAUAAUUUUUU____AGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA__AAAUCAACAAAA_ ........((((((...((((.......((((((((..((((..((((((((((........)))))))))))))).))))))))......))))..)))))). (-25.62 = -26.15 + 0.53)
| Location | 2,476,604 – 2,476,701 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.33 |
| Shannon entropy | 0.29492 |
| G+C content | 0.42156 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -24.96 |
| Energy contribution | -27.19 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.998568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2476604 97 - 23011544 -UUUUGUUGAUUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCU----AAAAAAUUAUACAAAAAACGGCAA -((((((((((((--(...(((((((((((((((((...((((......))))...))))..))).))))))))))----..))))))).))))))........ ( -37.90, z-score = -5.01, R) >droSim1.chr2L 2438607 97 - 22036055 -UUUUGUUGACUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCU----AAAAAAUUAUACAAAAAACGGCAA -((((((......--....(((((((((((((((((...((((......))))...))))..))).))))))))))----..........))))))........ ( -34.45, z-score = -3.76, R) >droSec1.super_5 650230 97 - 5866729 -UUUUGUUGACUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCU----AAAAAAUUAUACAAAAAACGGCAA -((((((......--....(((((((((((((((((...((((......))))...))))..))).))))))))))----..........))))))........ ( -34.45, z-score = -3.76, R) >droYak2.chr2L 2467727 104 - 22324452 UUUUUGUUGAUUUGCUGUGAGGUGCUGUUGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCCAAAAAAAAAAUUAUACAAAAAACGGCAA (((((((((((((....((.((((((((((((((((...((((......))))...))))..)))).))))))))))......)))))).)))))))....... ( -36.10, z-score = -2.95, R) >droEre2.scaffold_4929 2521044 96 - 26641161 -UUUUGUUGAUUU--UUUGAGGUGCUGUCGCUUGGGUACGACGUUAAUUUGUCACACCCAGCAGCAAGCAGCACCU----AAAAAAUUAUAC-AAAAACGGCAA -((((((((((((--(((.(((((((((.((((((((..((((......))))..)))))..)))..)))))))))----))))))))).))-))))....... ( -38.40, z-score = -5.68, R) >droAna3.scaffold_12913 218966 92 + 441482 -UUUUGUUGUU---------GAUGCUGGCAGUUGGGCGCGAUGUUAAUUUGUCACACCCAGCAGCA--CCACAAAAAAAUAAAUAAAAGGAAAGAAAAAGACAA -.((((((.((---------(.((...((.((((((.(.((((......)))).).)))))).)).--.)))))...))))))..................... ( -19.90, z-score = -0.62, R) >consensus _UUUUGUUGAUUU__UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCU____AAAAAAUUAUACAAAAAACGGCAA .((((((((((((......(((((((((((((((((...((((......))))...))))..))).)))))))))).......)))))).))))))........ (-24.96 = -27.19 + 2.23)
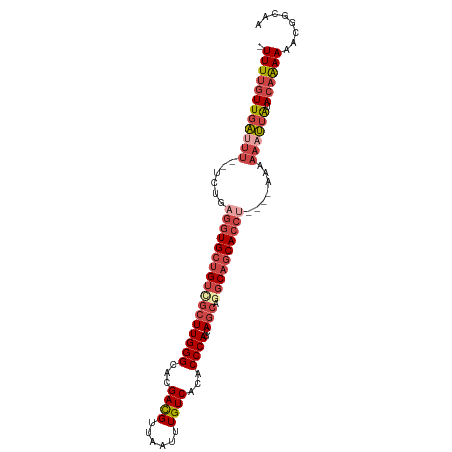
| Location | 2,476,625 – 2,476,732 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.06 |
| Shannon entropy | 0.17346 |
| G+C content | 0.51563 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -32.86 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2476625 107 + 23011544 UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAAUCAACAAAA-UCCUCAACGAAAUCGCGCGACGUCGAUACGA----- ...((((((((..((((..((((..((((........))))..)))))))).))))))))....--............-.......(((..((....))..)))......----- ( -35.70, z-score = -2.84, R) >droSim1.chr2L 2438628 112 + 22036055 UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAGUCAACAAAA-UCCUCAACGAAAUCGCGCGACGUCGAUACGACGUGC ...((((((((..((((..((((..((((........))))..)))))))).))))))))....--............-..............(((((.(((....))).))))) ( -40.00, z-score = -2.79, R) >droSec1.super_5 650251 112 + 5866729 UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAGUCAACAAAA-UCCUCAACGAAAUCGCGCGACGUCGAUACGACGUUC ...((((((((..((((..((((..((((........))))..)))))))).))))))))....--............-..................(((((((...))))))). ( -38.90, z-score = -2.81, R) >droYak2.chr2L 2467752 104 + 22324452 UUUGGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCAACAGCACCUCACAGCAAAUCAACAAAAAUCCUCAGCGAAAUCGCGCGACGUGC----------- ...((((((((..((((..((((..((((........))))..)))))))).)))))))).........................(((....))).........----------- ( -36.70, z-score = -2.32, R) >droEre2.scaffold_4929 2521064 112 + 26641161 UUUAGGUGCUGCUUGCUGCUGGGUGUGACAAAUUAACGUCGUACCCAAGCGACAGCACCUCAAA--AAAUCAACAAAA-UCCUCAACGAAAUCGCGCGACGUCGGUACGACGUGC ...((((((((.(((((..((((((((((........)))))))))))))))))))))))....--............-..............(((((.(((....))).))))) ( -40.60, z-score = -3.77, R) >consensus UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA__AAAUCAACAAAA_UCCUCAACGAAAUCGCGCGACGUCGAUACGACGU_C ...((((((((..((((..((((..((((........))))..)))))))).))))))))..........................(((..((....))..)))........... (-32.86 = -32.94 + 0.08)

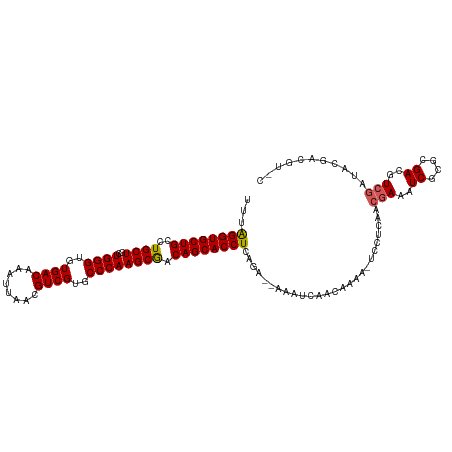
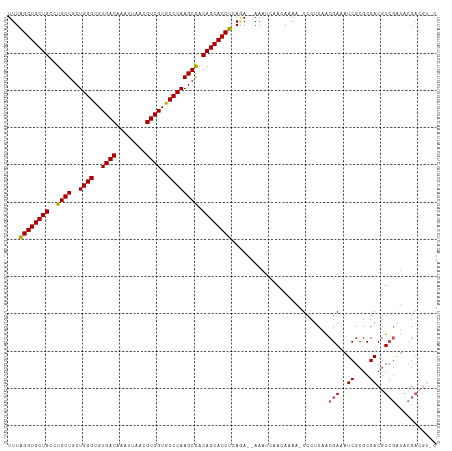
| Location | 2,476,625 – 2,476,732 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.06 |
| Shannon entropy | 0.17346 |
| G+C content | 0.51563 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -33.66 |
| Energy contribution | -33.78 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
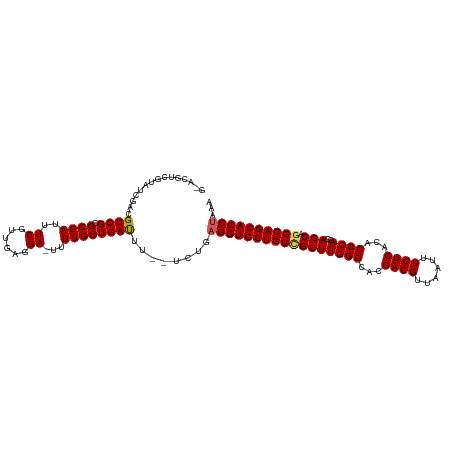
>dm3.chr2L 2476625 107 - 23011544 -----UCGUAUCGACGUCGCGCGAUUUCGUUGAGGA-UUUUGUUGAUUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA -----......(((.(((....))).)))..(((((-((.....)))))--))..(((((((((((((((((...((((......))))...))))..))).))))))))))... ( -38.20, z-score = -2.22, R) >droSim1.chr2L 2438628 112 - 22036055 GCACGUCGUAUCGACGUCGCGCGAUUUCGUUGAGGA-UUUUGUUGACUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA ((((((((...)))))).))((((..((......))-..))))......--....(((((((((((((((((...((((......))))...))))..))).))))))))))... ( -43.00, z-score = -2.60, R) >droSec1.super_5 650251 112 - 5866729 GAACGUCGUAUCGACGUCGCGCGAUUUCGUUGAGGA-UUUUGUUGACUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA (((.((((...))))((((.((((..((......))-..)))))))).)--))..(((((((((((((((((...((((......))))...))))..))).))))))))))... ( -41.40, z-score = -2.29, R) >droYak2.chr2L 2467752 104 - 22324452 -----------GCACGUCGCGCGAUUUCGCUGAGGAUUUUUGUUGAUUUGCUGUGAGGUGCUGUUGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCCAAA -----------(((.((((.((((.(((.....)))...)))))))).)))..((.((((((((((((((((...((((......))))...))))..)))).)))))))))).. ( -37.60, z-score = -1.24, R) >droEre2.scaffold_4929 2521064 112 - 26641161 GCACGUCGUACCGACGUCGCGCGAUUUCGUUGAGGA-UUUUGUUGAUUU--UUUGAGGUGCUGUCGCUUGGGUACGACGUUAAUUUGUCACACCCAGCAGCAAGCAGCACCUAAA ((((((((...)))))).))((((..((......))-..))))......--....(((((((((.((((((((..((((......))))..)))))..)))..)))))))))... ( -41.60, z-score = -2.98, R) >consensus G_ACGUCGUAUCGACGUCGCGCGAUUUCGUUGAGGA_UUUUGUUGAUUU__UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA ..............(.(((.(((....)))))).)....................(((((((((((((((((...((((......))))...))))..))).))))))))))... (-33.66 = -33.78 + 0.12)

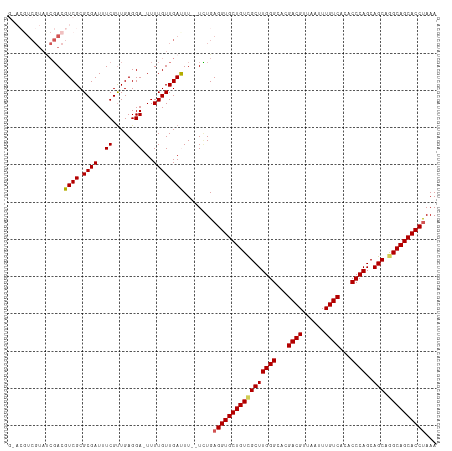
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:29 2011