| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,628,719 – 6,628,819 |
| Length | 100 |
| Max. P | 0.672144 |

| Location | 6,628,719 – 6,628,819 |
|---|---|
| Length | 100 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.53987 |
| G+C content | 0.52851 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -24.07 |
| Energy contribution | -23.15 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6628719 100 - 21146708 AGGGCUGCGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUAAGU--UCCUAGAAU---CUGGCAAGUCUAGG-------- .((((.(..((((((.(((((((..((...))..))))))).))))..))..).)))).(((((...(((.......--)))......---))))).........-------- ( -37.10, z-score = -0.74, R) >anoGam1.chr2L 9908436 100 - 48795086 AGGGCUGCGCGCAGAUGAUGUGCAAACGGUGCAAGCACGUCUUCUGCUGGUACUGUCUCGCCAGUCUGGACGUAAGUGUUCUGCAU-----CUUAUCGUUGUGCA-------- ......(((((..(((((.((((((((((((....)))((((...((((((........))))))..)))).....)))).)))))-----.)))))..))))).-------- ( -35.00, z-score = -0.44, R) >droGri2.scaffold_15245 5461751 99 - 18325388 AGGGCUGUGCCCAGAUGAUGUGCAAGCGUUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUGAGUAGACUCUGCAG---UUGACAGCUUG----------- .((((.(((((((((.(((((((..((...))..))))))).))))..))))).))))(((..(((....((((((....))).))).---..))).)))..----------- ( -37.30, z-score = -0.85, R) >droMoj3.scaffold_6496 10272542 92 - 26866924 AGGGCUGCGCCCAGAUGAUGUGCAAGCGGUGCAAGCACGUCUUCUGCUGGUACUGUCUGGCCAGUCUCGAUGUGAGUGUUGUCUGUUA---UUUG------------------ .((((...))))............(((((.(((..((((((....((((((........))))))...))))))..)))...))))).---....------------------ ( -30.00, z-score = -0.20, R) >droVir3.scaffold_12875 16637214 97 - 20611582 AGGGAUGUGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGUCUGGCCAGCCUGGAUGUGAGUA-UAAUAG--------UGAGAGUUAAUUG------- ..(((((((((((((.(((((((..((...))..))))))).))))..))))).))))....(((((.(.(((.....-..))).--------).)).))).....------- ( -26.90, z-score = 0.52, R) >droWil1.scaffold_180699 697332 100 + 2593675 AGGGCUGUGCCCAAAUGAUGUGCAAAAGGUGCAAGCAUGUAUUCUGCUGGUAUUGCCUAGCCAGCUUGGAUGUGAGUAUUAAUAGA-----CUUGGAAGUGAUUU-------- .((((...))))..(((...((((.....))))..))).(((((.((((((........))))))..))))).((((........)-----)))...........-------- ( -24.70, z-score = 0.58, R) >droPer1.super_4 749301 113 - 7162766 AGGGCUGUGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUUUGCUGGUACUGCCUGGCCAGUCUGGAUGUGAGUGGCGGCGAGAUGUGUUCUCGAGUCGGCGAGUAGCUA .((((.(((((((((.(((((((..((...))..))))))).))))..))))).)))).((((.((.......)).))))..(((((.....)))))....(((.....))). ( -45.40, z-score = -0.78, R) >dp4.chr3 3765468 113 + 19779522 AGGGCUGUGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUUUGCUGGUACUGCCUGGCCAGUCUGGAUGUGAGUGGCUGCGAGAUGUGUUCUCGAGUCGGCGAGUAGCUA .((((.(((((((((.(((((((..((...))..))))))).))))..))))).))))(((((.((.......)).))))).(((((.....)))))....(((.....))). ( -46.20, z-score = -1.19, R) >droAna3.scaffold_13266 18120657 94 - 19884421 AGGGCUGUGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGUCUGGCCAGUUUGGAUGUGAGU-UCUGGAAUAU---UUUAGCC--------------- ..(((((...(((((.....((((.....))))..(((((((...((((((........))))))..)))))))...-))))).....---..)))))--------------- ( -32.00, z-score = -0.74, R) >droEre2.scaffold_4845 3440300 101 + 22589142 AGGGCUGCGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUAAGU-GCCACAAAAU---CCCACAAAUGGUGG-------- .((((...)))).(((..((((...(((((......((((((...((((((........))))))..)))))).)))-))))))..))---)((((.....))))-------- ( -35.00, z-score = -0.09, R) >droYak2.chr2R 18608793 98 + 21139217 AGGGCUGCGCCCAAAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUAAGU--CCAUUGAAU---CUCACAAAUUAU---------- .((((((((.((((..(((((((..((...))..)))))))....((((((........)))))))))).))).)))--)).......---............---------- ( -31.30, z-score = -0.78, R) >droSec1.super_1 4209046 99 - 14215200 AGGGCUGCGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUAAGU--CCCUAG-AU---CUUACAAGUCUAGG-------- .((((((((.(((((.(((((((..((...))..))))))).)))((((((........))))))..)).))).)))--))((((-((---.......)))))).-------- ( -36.50, z-score = -1.17, R) >droSim1.chr2R 5235316 99 - 19596830 AGGGCUGCGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUAAGU--CCCUAG-AU---CUUACAUGUCUAGA-------- .((((((((.(((((.(((((((..((...))..))))))).)))((((((........))))))..)).))).)))--))((((-((---.......)))))).-------- ( -36.10, z-score = -1.20, R) >triCas2.ChLG3 19520110 98 - 32080666 AAGGCUGUGCGCAAAUGAUGUGUAAACGGUGCAAACACGUCUUUUGCUGGUACUGUCUCGCCAGUCUCGAUGUAA----UAACAACGC---CCUAUUAGUGUUUG-------- ...((((((((((.....)))))..))))).(((((((.......((((((........))))))..((.(((..----..))).)).---.......)))))))-------- ( -24.10, z-score = -0.18, R) >consensus AGGGCUGCGCCCAGAUGAUGUGCAAGCGCUGCAAGCACGUCUUCUGCUGGUACUGCCUGGCCAGUUUGGAUGUAAGU__CCACAGAAU___CUUACAAGUCUA_G________ .((((((.(((((((.(((((((...........))))))).))))..((.....)).))))))))).............................................. (-24.07 = -23.15 + -0.92)

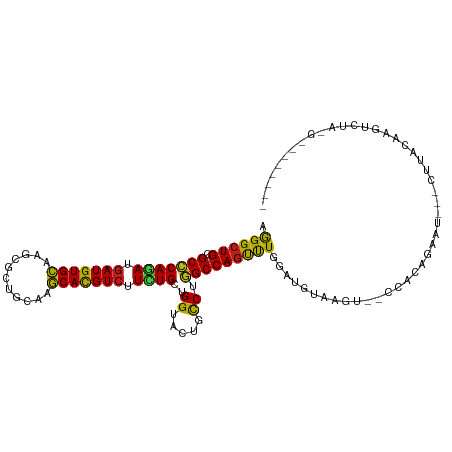
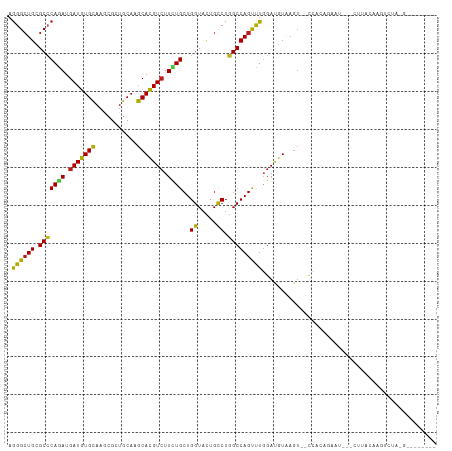
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:13 2011