
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,604,916 – 6,605,050 |
| Length | 134 |
| Max. P | 0.997226 |

| Location | 6,604,916 – 6,605,050 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 164 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Shannon entropy | 0.44922 |
| G+C content | 0.46511 |
| Mean single sequence MFE | -48.98 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.52 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.997226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
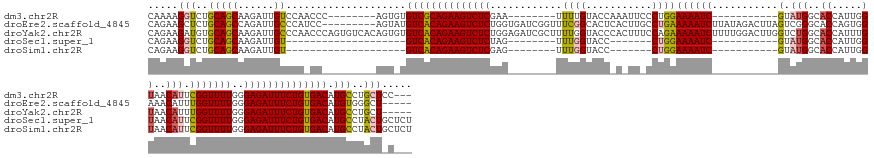
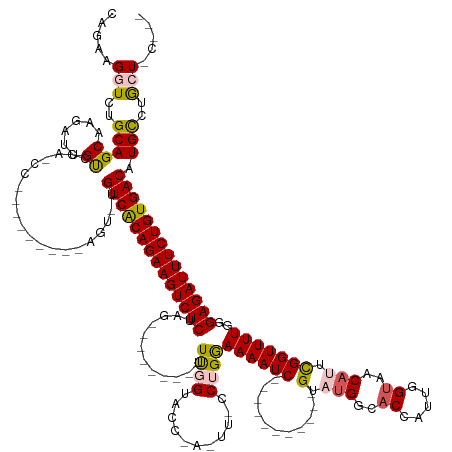
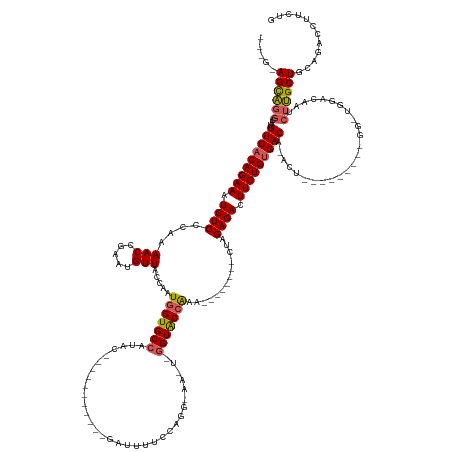
>dm3.chr2R 6604916 134 + 21146708 CAAAAGGUCUGCAGCAAGAUUGUCCAACCC--------AGUGUGUCGCAGAAGUCUCGAA--------UUUUGUACCAAAUUCCCUGGAAAAUC-----------GUAUGGCACCAUUGGUAACAUUCGGUUUUGGGAGAUUUCUGUGACAUGCCUGCUCC--- .....((...((((...(......).....--------.(((((((((((((((((((((--------(((......))))))((...((((((-----------(.(((..(((...)))..))).))))))))))))))))))))))))))))))).))--- ( -45.60, z-score = -3.34, R) >droEre2.scaffold_4845 3416768 150 - 22589142 CAGAAGCUCUGCAGCCAGAUUGCCCAUCC---------AGUAUGUCACAGAAGUCUCUGGUGAUCGGUUUCGGCACUCACUUGCCUGAAAAAUCUUAUAGACUUAGUCGGGCACCAGUGGAAACAUUUGGUUUUGGGAGAUUUCUGUGACAUGUGGGCU----- .......((((....))))..((((((..---------...(((((((((((((((((((((.(((....))))))).....((((((....((.....)).....))))))(((((((....)).)))))....))))))))))))))))))))))).----- ( -59.40, z-score = -3.78, R) >droYak2.chr2R 18579400 159 - 21139217 CAGAAGAUGUGCAGCAAGAUUGCCCAACCCAGUGUCACAGUGUGUCACAGAAGUCUCUGGAGAUCGCUUUUGGUACCCACUUUCCAGAAAAAUCUUUUGGACUUGGUCUGGCACCAUUUGUAACAUUUGGUUUUGGGAGAUUUCUGUGACAUGCCUGCU----- (((....(((((((((....))).........)).))))(((((((((((((((((((.((((((((...((((.((.(((.(((((((.....)))))))...)))..)).))))...)).......)))))).))))))))))))))))))))))..----- ( -54.91, z-score = -2.31, R) >droSec1.super_1 4185330 118 + 14215200 CAGAAGGUCUGCAGCAAGAUUGU--------------------GUCACAGAAGUCUCUAG--------UUUGGUACC-------CUGGAAAAUC-----------GUAUGGCACCAUUGGUAACAUUCGGUUUUGGGAGAUUUCUGUGACAUGCCUACUGCUCU (((.((((((......))...((--------------------(((((((((((((((..--------.(..(....-------)..)((((((-----------(.(((..(((...)))..))).))))))).))))))))))))))))))))).))).... ( -43.00, z-score = -3.20, R) >droSim1.chr2R 5211549 118 + 19596830 CAGAAGGUCUGCAGCAAGAUUGU--------------------GUCACAGAAGUCUCGAG--------UUUGGUACC-------CUGGAAAAUC-----------GUAUGGCACCAUUGGUAACAUUCGGUUUUGGGAGAUUUCUGUGACAUGCCUACUGCUCU (((.((((((......))...((--------------------((((((((((((((...--------.(..(....-------)..)((((((-----------(.(((..(((...)))..))).)))))))..)))))))))))))))))))).))).... ( -42.00, z-score = -2.82, R) >consensus CAGAAGGUCUGCAGCAAGAUUGUCCA_CC_________AGU_UGUCACAGAAGUCUCUAG________UUUGGUACC_A_UU_CCUGGAAAAUC___________GUAUGGCACCAUUGGUAACAUUCGGUUUUGGGAGAUUUCUGUGACAUGCCUGCU_C___ .....(((..(((((......))....................((((((((((((((................................................(.....).(((.....(((.....))).))))))))))))))))).)))..)))..... (-25.92 = -25.52 + -0.40)
| Location | 6,604,916 – 6,605,050 |
|---|---|
| Length | 134 |
| Sequences | 5 |
| Columns | 164 |
| Reading direction | reverse |
| Mean pairwise identity | 71.72 |
| Shannon entropy | 0.44922 |
| G+C content | 0.46511 |
| Mean single sequence MFE | -42.14 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
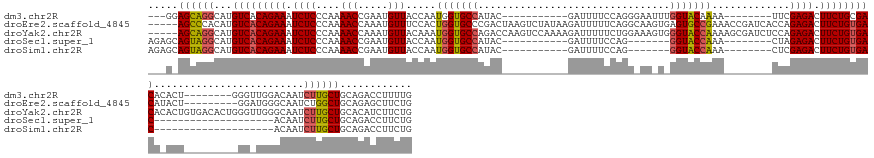
>dm3.chr2R 6604916 134 - 21146708 ---GGAGCAGGCAUGUCACAGAAAUCUCCCAAAACCGAAUGUUACCAAUGGUGCCAUAC-----------GAUUUUCCAGGGAAUUUGGUACAAAA--------UUCGAGACUUCUGCGACACACU--------GGGUUGGACAAUCUUGCUGCAGACCUUUUG ---(.((((((..((((.(((((.((((((((((.((.(((.((((...)))).))).)-----------).))))...))((((((......)))--------))))))).)))))((((.....--------..))))))))..)))))).).......... ( -37.20, z-score = -1.21, R) >droEre2.scaffold_4845 3416768 150 + 22589142 -----AGCCCACAUGUCACAGAAAUCUCCCAAAACCAAAUGUUUCCACUGGUGCCCGACUAAGUCUAUAAGAUUUUUCAGGCAAGUGAGUGCCGAAACCGAUCACCAGAGACUUCUGUGACAUACU---------GGAUGGGCAAUCUGGCUGCAGAGCUUCUG -----.(((((((((((((((((.((((............(((((((((..((((.((..(((((.....))))).)).))))....))))..))))).........)))).)))))))))))...---------.).)))))..((((....))))....... ( -47.40, z-score = -2.53, R) >droYak2.chr2R 18579400 159 + 21139217 -----AGCAGGCAUGUCACAGAAAUCUCCCAAAACCAAAUGUUACAAAUGGUGCCAGACCAAGUCCAAAAGAUUUUUCUGGAAAGUGGGUACCAAAAGCGAUCUCCAGAGACUUCUGUGACACACUGUGACACUGGGUUGGGCAAUCUUGCUGCACAUCUUCUG -----.((((...((((((((((.((((....(((.....))).....(((((((..((....((((...((....))))))..)).))))))).............)))).))))))))))..))))..((..((((((((((....)))).)).))))..)) ( -44.40, z-score = -0.47, R) >droSec1.super_1 4185330 118 - 14215200 AGAGCAGUAGGCAUGUCACAGAAAUCUCCCAAAACCGAAUGUUACCAAUGGUGCCAUAC-----------GAUUUUCCAG-------GGUACCAAA--------CUAGAGACUUCUGUGAC--------------------ACAAUCUUGCUGCAGACCUUCUG ...((((((((..((((((((((.((((....(((.....))).....(((((((....-----------(.....)...-------)))))))..--------...)))).)))))))))--------------------)....)))))))).......... ( -40.80, z-score = -4.52, R) >droSim1.chr2R 5211549 118 - 19596830 AGAGCAGUAGGCAUGUCACAGAAAUCUCCCAAAACCGAAUGUUACCAAUGGUGCCAUAC-----------GAUUUUCCAG-------GGUACCAAA--------CUCGAGACUUCUGUGAC--------------------ACAAUCUUGCUGCAGACCUUCUG ...((((((((..((((((((((.((((....(((.....))).....(((((((....-----------(.....)...-------)))))))..--------...)))).)))))))))--------------------)....)))))))).......... ( -40.90, z-score = -4.59, R) >consensus ___G_AGCAGGCAUGUCACAGAAAUCUCCCAAAACCGAAUGUUACCAAUGGUGCCAUAC___________GAUUUUCCAGG_AA_U_GGUACCAAA________CUAGAGACUUCUGUGACA_ACU_________GG_UGGACAAUCUUGCUGCAGACCUUCUG .....((((((..((((((((((.((((....(((.....))).....(((((((................................))))))).............)))).))))))))))........................))))))............ (-24.84 = -25.36 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:11 2011