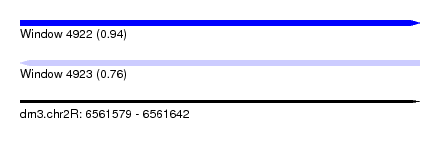
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,561,579 – 6,561,642 |
| Length | 63 |
| Max. P | 0.940266 |

| Location | 6,561,579 – 6,561,642 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Shannon entropy | 0.48644 |
| G+C content | 0.50625 |
| Mean single sequence MFE | -18.76 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.26 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
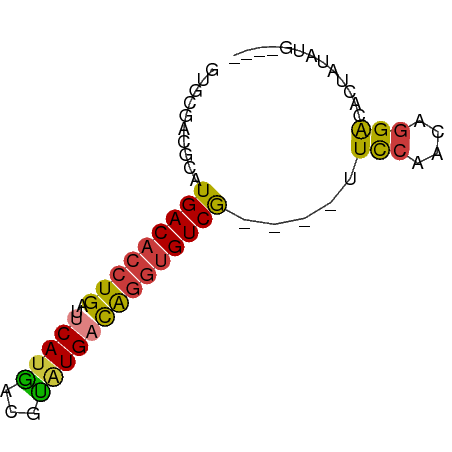
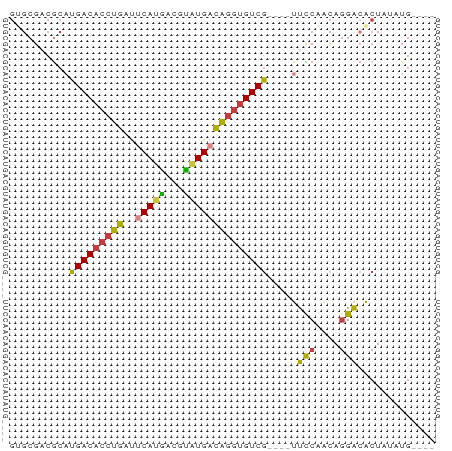
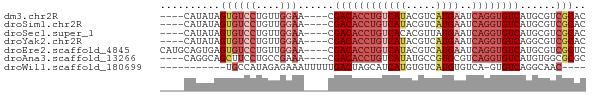
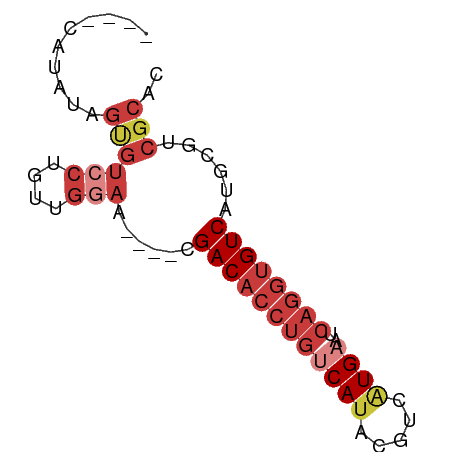
>dm3.chr2R 6561579 63 + 21146708 GUGCGACGCAUGACACCUGAUUCAUGACGUAUGACAGGUGUCG----UUCCAACAGGACACUAUAUG---- (((......((((((((((..(((((...))))))))))))))----)(((....))))))......---- ( -19.80, z-score = -1.82, R) >droSim1.chr2R 5166388 63 + 19596830 GUGCGACGCAUGACACCUGAUUCAUGACGUAUGACAGGUGUCG----UUCCAACAGGACACUAUAUG---- (((......((((((((((..(((((...))))))))))))))----)(((....))))))......---- ( -19.80, z-score = -1.82, R) >droSec1.super_1 4141922 63 + 14215200 GUGCGACGCAUGACACCUGAUUCAUAACGUGUGACAGGUGUCG----UUCCAACAGGACACUAUAUG---- (((......((((((((((..((((.....)))))))))))))----)(((....))))))......---- ( -20.20, z-score = -1.78, R) >droYak2.chr2R 18535894 63 - 21139217 GUGCGACGCCUGACACCUGAUUCAUGACGUAUGACAGGUGUCG----UUCCAACAGGACACUAUAUG---- (((........((((((((..(((((...))))))))))))).----.(((....))))))......---- ( -18.80, z-score = -1.58, R) >droEre2.scaffold_4845 3374295 67 - 22589142 GAGCGACGCAUGACACCUGAUUCAUGACGUAUGACAGGUGUCG----UUCCAACAGGACACUCACUGCAUG (((.(....((((((((((..(((((...))))))))))))))----)(((....)))).)))........ ( -21.60, z-score = -1.86, R) >droAna3.scaffold_13266 18050533 63 + 19884421 GCGCGCCACAUGACACCUGACGCACGGCAUAUGACAGGUGUCG----UUUCGGCAGGAAGCUGCCUG---- .........((((((((((...((.......)).)))))))))----)...(((((....)))))..---- ( -23.50, z-score = -1.32, R) >droWil1.scaffold_180699 631101 55 - 2593675 ----GUUGCCUGACAC-UGACACAUGACACAUGAUGCUAGUCAAAAAUUUCUCUAUGGCA----------- ----..((((((((..-.(.((((((...)))).)))..)))).............))))----------- ( -7.61, z-score = 0.49, R) >consensus GUGCGACGCAUGACACCUGAUUCAUGACGUAUGACAGGUGUCG____UUCCAACAGGACACUAUAUG____ ...........((((((((..(((((...)))))))))))))......(((....)))............. (-13.17 = -13.26 + 0.09)

| Location | 6,561,579 – 6,561,642 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 74.69 |
| Shannon entropy | 0.48644 |
| G+C content | 0.50625 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -12.22 |
| Energy contribution | -14.06 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6561579 63 - 21146708 ----CAUAUAGUGUCCUGUUGGAA----CGACACCUGUCAUACGUCAUGAAUCAGGUGUCAUGCGUCGCAC ----......((((((....))).----.((((((((((((.....))))..))))))))......))).. ( -19.90, z-score = -1.49, R) >droSim1.chr2R 5166388 63 - 19596830 ----CAUAUAGUGUCCUGUUGGAA----CGACACCUGUCAUACGUCAUGAAUCAGGUGUCAUGCGUCGCAC ----......((((((....))).----.((((((((((((.....))))..))))))))......))).. ( -19.90, z-score = -1.49, R) >droSec1.super_1 4141922 63 - 14215200 ----CAUAUAGUGUCCUGUUGGAA----CGACACCUGUCACACGUUAUGAAUCAGGUGUCAUGCGUCGCAC ----......((((((....))).----.(((((((((((.......)))..))))))))......))).. ( -18.30, z-score = -0.85, R) >droYak2.chr2R 18535894 63 + 21139217 ----CAUAUAGUGUCCUGUUGGAA----CGACACCUGUCAUACGUCAUGAAUCAGGUGUCAGGCGUCGCAC ----......((((((....))).----.((((((((((((.....))))..))))))))......))).. ( -19.90, z-score = -1.45, R) >droEre2.scaffold_4845 3374295 67 + 22589142 CAUGCAGUGAGUGUCCUGUUGGAA----CGACACCUGUCAUACGUCAUGAAUCAGGUGUCAUGCGUCGCUC .(((((....((.(((....))))----)((((((((((((.....))))..)))))))).)))))..... ( -22.90, z-score = -1.41, R) >droAna3.scaffold_13266 18050533 63 - 19884421 ----CAGGCAGCUUCCUGCCGAAA----CGACACCUGUCAUAUGCCGUGCGUCAGGUGUCAUGUGGCGCGC ----..(((((....)))))....----.((((((((....((((...))))))))))))........... ( -22.70, z-score = -0.64, R) >droWil1.scaffold_180699 631101 55 + 2593675 -----------UGCCAUAGAGAAAUUUUUGACUAGCAUCAUGUGUCAUGUGUCA-GUGUCAGGCAAC---- -----------((((..(((....))).((((..(((.((((...)))))))..-..))))))))..---- ( -12.40, z-score = -0.55, R) >consensus ____CAUAUAGUGUCCUGUUGGAA____CGACACCUGUCAUACGUCAUGAAUCAGGUGUCAUGCGUCGCAC ..........((((((....)))......((((((((((((.....))))..))))))))......))).. (-12.22 = -14.06 + 1.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:13:00 2011