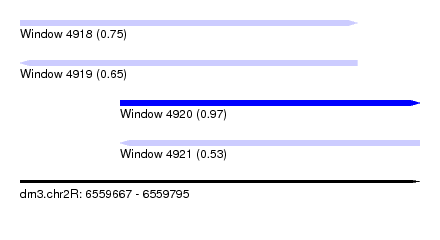
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,559,667 – 6,559,795 |
| Length | 128 |
| Max. P | 0.974861 |

| Location | 6,559,667 – 6,559,775 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Shannon entropy | 0.37626 |
| G+C content | 0.48749 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -16.73 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6559667 108 + 21146708 UAUUCGGAAGUAUUCGGACCACGUCAGCUGGCCAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCC--AACACCUUCCUGUAAGCUAUAAAUUCCG-CCGACACAUA ....((((((((((.((.((((....).))))).)))))(((.((((((((((..--(((...)))))))))--)))).)))...............)))))-.......... ( -33.20, z-score = -2.28, R) >droSim1.chr2R 5164504 108 + 19596830 UGUUCGGAAGUAUUCGGACCACGUCAGCUGGCCAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCC--AACACCUUCCUGUAAGCUAUAAAUUCCG-CCGACACAUA ((((((((((((((.((.((((....).))))).)))))(((.((((((((((..--(((...)))))))))--)))).)))...............)))))-..)))).... ( -35.00, z-score = -2.43, R) >droSec1.super_1 4140088 108 + 14215200 UGUUCGGAAGUAUUCGGACCACGUCAGCUGGCCAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCC--AACACCUUCCUGUAAGCUAUAAAUUCCG-CCGACACAUC ((((((((((((((.((.((((....).))))).)))))(((.((((((((((..--(((...)))))))))--)))).)))...............)))))-..)))).... ( -35.00, z-score = -2.41, R) >droYak2.chr2R 18531670 108 - 21139217 UGUUCGGAAGUAUUCGGACCACGUCAGCUGGCCAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCC--AACACCUUCCUGUAAGCUAUAAAUUCCG-CCGACACAAA ((((((((((((((.((.((((....).))))).)))))(((.((((((((((..--(((...)))))))))--)))).)))...............)))))-..)))).... ( -35.00, z-score = -2.44, R) >droEre2.scaffold_4845 3372513 110 - 22589142 CGUUCGGAAGUAUUCGGCCCACGUCAGCUGGCCAAAUGCGAGUUGUUGGCCAAACACGAGGAUGUCUUGGCC--AACACCUUCCUGUAAGCUAUAAAUUCCG-UCGACAAAUG ((..((((((((((.((((..(....)..)))).)))))(((.(((((((((((((......))).))))))--)))).)))...............)))))-.))....... ( -37.60, z-score = -2.91, R) >droGri2.scaffold_15245 5377371 101 + 18325388 UGUGUGGCAUCCUUUGGGCCAUGUC-----------UACGUGUUUACAGUAACACA-UGCAAUUUGUUGGCCCUAUUUGCUGCCAAAAUGUUGUAGUUUCUGGUUAACAAAUG ((((((((((((....))..))).)-----------)))(((((......))))).-.)))(((((((((((......(((((.........)))))....))))))))))). ( -26.40, z-score = -0.57, R) >consensus UGUUCGGAAGUAUUCGGACCACGUCAGCUGGCCAAAUGCGAGUUGUUGGCCAAAC__GACGAUGUCUUGGCC__AACACCUUCCUGUAAGCUAUAAAUUCCG_CCGACACAUA .....(((((.....((.(((((((...(((((((..........))))))).....))))......))))).......)))))............................. (-16.73 = -17.70 + 0.97)
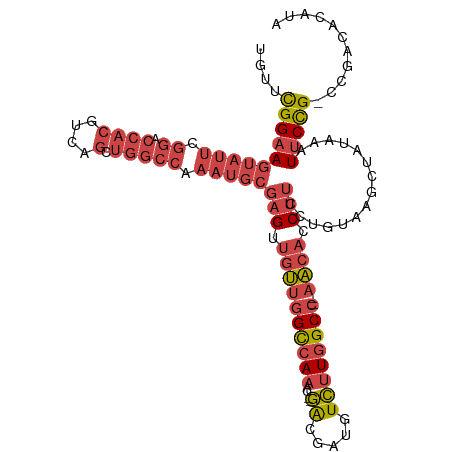
| Location | 6,559,667 – 6,559,775 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Shannon entropy | 0.37626 |
| G+C content | 0.48749 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -19.05 |
| Energy contribution | -20.55 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6559667 108 - 21146708 UAUGUGUCGG-CGGAAUUUAUAGCUUACAGGAAGGUGUU--GGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUGGCCAGCUGACGUGGUCCGAAUACUUCCGAAUA ....(((.((-(..........))).)))(((((...((--(((((......((((--(.((((((((..........)))))))).)))))))).))))...)))))..... ( -36.60, z-score = -2.00, R) >droSim1.chr2R 5164504 108 - 19596830 UAUGUGUCGG-CGGAAUUUAUAGCUUACAGGAAGGUGUU--GGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUGGCCAGCUGACGUGGUCCGAAUACUUCCGAACA ....(((.((-(..........))).)))(((((...((--(((((......((((--(.((((((((..........)))))))).)))))))).))))...)))))..... ( -36.60, z-score = -1.87, R) >droSec1.super_1 4140088 108 - 14215200 GAUGUGUCGG-CGGAAUUUAUAGCUUACAGGAAGGUGUU--GGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUGGCCAGCUGACGUGGUCCGAAUACUUCCGAACA ....(((.((-(..........))).)))(((((...((--(((((......((((--(.((((((((..........)))))))).)))))))).))))...)))))..... ( -36.60, z-score = -1.67, R) >droYak2.chr2R 18531670 108 + 21139217 UUUGUGUCGG-CGGAAUUUAUAGCUUACAGGAAGGUGUU--GGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUGGCCAGCUGACGUGGUCCGAAUACUUCCGAACA ....(((.((-(..........))).)))(((((...((--(((((......((((--(.((((((((..........)))))))).)))))))).))))...)))))..... ( -36.60, z-score = -1.84, R) >droEre2.scaffold_4845 3372513 110 + 22589142 CAUUUGUCGA-CGGAAUUUAUAGCUUACAGGAAGGUGUU--GGCCAAGACAUCCUCGUGUUUGGCCAACAACUCGCAUUUGGCCAGCUGACGUGGGCCGAAUACUUCCGAACG ..((((....-))))..............(((((.((((--(((((.(((((....))))))))))))))......((((((((.((....)).)))))))).)))))..... ( -38.50, z-score = -2.70, R) >droGri2.scaffold_15245 5377371 101 - 18325388 CAUUUGUUAACCAGAAACUACAACAUUUUGGCAGCAAAUAGGGCCAACAAAUUGCA-UGUGUUACUGUAAACACGUA-----------GACAUGGCCCAAAGGAUGCCACACA .(((((((..((((((.........)))))).))))))).((((((......((((-((((((......))))))).-----------).))))))))............... ( -24.80, z-score = -1.02, R) >consensus UAUGUGUCGG_CGGAAUUUAUAGCUUACAGGAAGGUGUU__GGCCAAGACAUCGUC__GUUUGGCCAACAACUCGCAUUUGGCCAGCUGACGUGGUCCGAAUACUUCCGAACA .............................(((((.......(((((......((((....((((((((..........))))))))..)))))))))......)))))..... (-19.05 = -20.55 + 1.50)
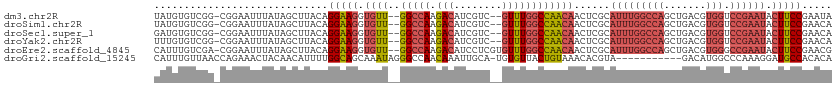
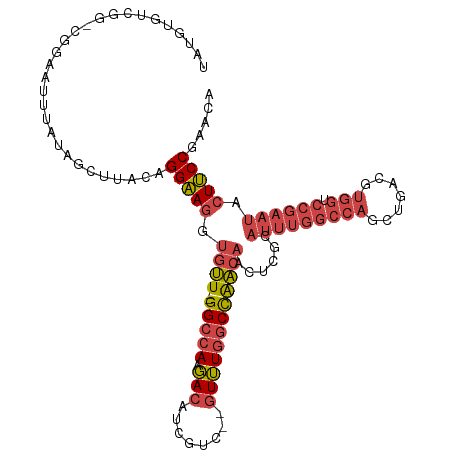
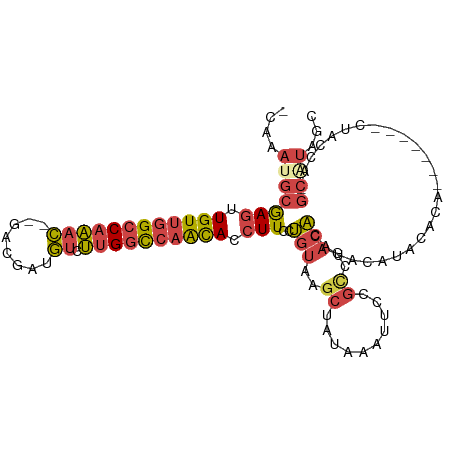
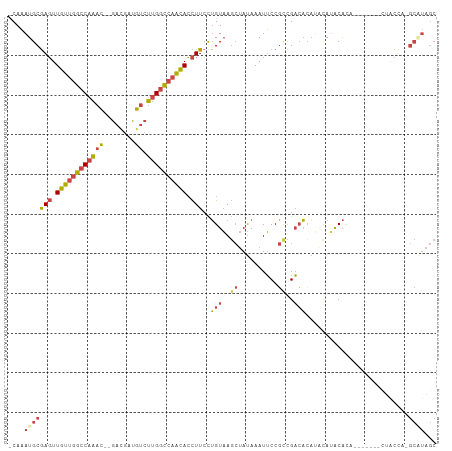
| Location | 6,559,699 – 6,559,795 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.52020 |
| G+C content | 0.46030 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -13.09 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6559699 96 + 21146708 -CAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCCAACACCUUCCUGUAAGCUAUAAAUUCCGCCGACACAUACAUACAU--------CUACCA-GCGUAGC -...(((((((.((((((((((..--(((...))))))))))))).)))..(((..((..........))..)))...........--------......-))))... ( -23.90, z-score = -1.98, R) >droSim1.chr2R 5164536 105 + 19596830 -CAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCCAACACCUUCCUGUAAGCUAUAAAUUCCGCCGACACAUAUAUACAUACAUACAUCUACCAAGCAUAGC -...(((((((.((((((((((..--(((...))))))))))))).)))..(((..((..........))..)))..........................))))... ( -24.30, z-score = -2.42, R) >droSec1.super_1 4140120 97 + 14215200 -CAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCCAACACCUUCCUGUAAGCUAUAAAUUCCGCCGACACAUCUAUACAU--------CUACCAAGCAUAGC -...(((((((.((((((((((..--(((...))))))))))))).)))..(((..((..........))..)))...........--------.......))))... ( -24.30, z-score = -2.58, R) >droYak2.chr2R 18531702 98 - 21139217 -CAAAUGCGAGUUGUUGGCCAAAC--GACGAUGUCUUGGCCAACACCUUCCUGUAAGCUAUAAAUUCCGCCGACACAAACAUACACA-------UCUCCUUGCAUAGC -...(((((((.((((((((((..--(((...)))))))))))))......(((..((..........))..)))............-------....)))))))... ( -26.10, z-score = -3.14, R) >droEre2.scaffold_4845 3372545 100 - 22589142 -CAAAUGCGAGUUGUUGGCCAAACACGAGGAUGUCUUGGCCAACACCUUCCUGUAAGCUAUAAAUUCCGUCGACAAAUGCACACACA------UCUACUU-GCUUAGC -.....(((((((((((((((((((......))).))))))))))......(((..((..........))..)))............------...))))-))..... ( -24.20, z-score = -1.33, R) >droGri2.scaffold_15245 5377411 82 + 18325388 ACACAUGCAAUUUGUUGGCCCUAUUUGCUGCCAAAAUGUU--GUAGUUUCUGGUUA---ACAAAUGCCUCUGCACCUGCCAAGCGCU--------------------- .........(((((((((((......(((((.........--)))))....)))))---))))))((((..((....))..)).)).--------------------- ( -19.10, z-score = -0.15, R) >consensus _CAAAUGCGAGUUGUUGGCCAAAC__GACGAUGUCUUGGCCAACACCUUCCUGUAAGCUAUAAAUUCCGCCGACACAUACAUACACA_______CUACCA_GCAUAGC ....(((((((.((((((((((.............)))))))))).)))..(((..((..........))..)))..........................))))... (-13.09 = -14.12 + 1.03)
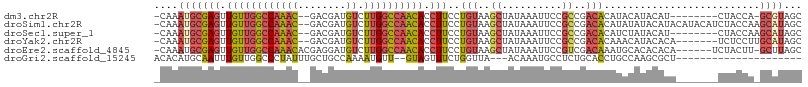
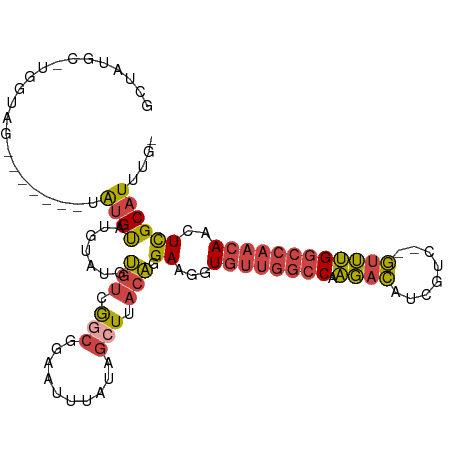
| Location | 6,559,699 – 6,559,795 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.52020 |
| G+C content | 0.46030 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.526515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
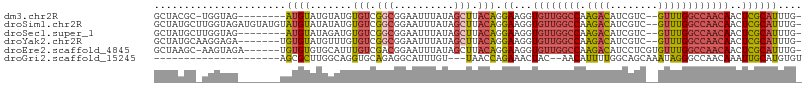
>dm3.chr2R 6559699 96 - 21146708 GCUACGC-UGGUAG--------AUGUAUGUAUGUGUCGGCGGAAUUUAUAGCUUACAGGAAGGUGUUGGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUG- ((..(((-((..(.--------(((....))).)..))))).................((...(((((((((.(((...)))--...)))))))))..)))).....- ( -30.10, z-score = -1.48, R) >droSim1.chr2R 5164536 105 - 19596830 GCUAUGCUUGGUAGAUGUAUGUAUGUAUAUAUGUGUCGGCGGAAUUUAUAGCUUACAGGAAGGUGUUGGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUG- ((((((.((.((.((..((((((....))))))..)).)).))...))))))......(..(((((((((((.(((...)))--...)))))))).)))..).....- ( -32.60, z-score = -1.84, R) >droSec1.super_1 4140120 97 - 14215200 GCUAUGCUUGGUAG--------AUGUAUAGAUGUGUCGGCGGAAUUUAUAGCUUACAGGAAGGUGUUGGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUG- ((((((.((.((.(--------(..((....))..)).)).))...))))))......(..(((((((((((.(((...)))--...)))))))).)))..).....- ( -27.80, z-score = -0.96, R) >droYak2.chr2R 18531702 98 + 21139217 GCUAUGCAAGGAGA-------UGUGUAUGUUUGUGUCGGCGGAAUUUAUAGCUUACAGGAAGGUGUUGGCCAAGACAUCGUC--GUUUGGCCAACAACUCGCAUUUG- ...........(((-------((((.((.(((.(((.(((..........))).))).))).))((((((((.(((...)))--...))))))))....))))))).- ( -29.90, z-score = -1.40, R) >droEre2.scaffold_4845 3372545 100 + 22589142 GCUAAGC-AAGUAGA------UGUGUGUGCAUUUGUCGACGGAAUUUAUAGCUUACAGGAAGGUGUUGGCCAAGACAUCCUCGUGUUUGGCCAACAACUCGCAUUUG- (((((((-...((((------(((....)))))))((....)).......)))))...((...(((((((((.(((((....))))))))))))))..)))).....- ( -29.20, z-score = -1.18, R) >droGri2.scaffold_15245 5377411 82 - 18325388 ---------------------AGCGCUUGGCAGGUGCAGAGGCAUUUGU---UAACCAGAAACUAC--AACAUUUUGGCAGCAAAUAGGGCCAACAAAUUGCAUGUGU ---------------------........(((.((((((.(((((((((---(..((((((.....--....)))))).)))))))...)))......)))))).))) ( -19.20, z-score = 0.77, R) >consensus GCUAUGC_UGGUAG_______UAUGUAUGUAUGUGUCGGCGGAAUUUAUAGCUUACAGGAAGGUGUUGGCCAAGACAUCGUC__GUUUGGCCAACAACUCGCAUUUG_ ......................((((.......(((.(((..........))).))).((...((((((((.((((........))))))))))))..)))))).... (-15.17 = -15.57 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:58 2011