| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,542,307 – 6,542,409 |
| Length | 102 |
| Max. P | 0.941412 |

| Location | 6,542,307 – 6,542,409 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.46096 |
| G+C content | 0.53825 |
| Mean single sequence MFE | -37.56 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.42 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
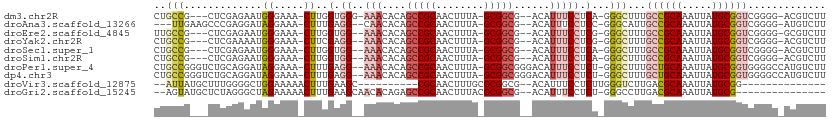
>dm3.chr2R 6542307 102 + 21146708 CUGCCG---CUCGAGAAUGGGAAA-CUUGGUGGG-AAACACAGCCGCAACUUUA-GCGGCG--ACAUUUCCUCA-GGGCUUUGCCGCAAAUUAUGCGGUCGGGG-ACGUCUU ..((((---((.(((...((....-)).(((.(.-...)...)))....))).)-)))))(--((....((...-.))((..((((((.....))))))..)).-..))).. ( -37.60, z-score = -1.70, R) >droAna3.scaffold_13266 18031824 101 + 19884421 ---UUGAAGCCCGAGGAUAGGAAA-CUUUGAGG--CAACACAGCCGCAACUUUA-GCGGCG--ACAUUUCCUCC-GGGCAUUGCCGCAAAUUAUGCGGUCGGGG-AUGUCUU ---.....((((((((........-))))).))--)......(((((.......-)))))(--((((..((.((-.(((...)))(((.....)))))..))..-))))).. ( -39.00, z-score = -2.70, R) >droEre2.scaffold_4845 3362175 101 - 22589142 UUGCCG---CUCGAGAAUGGGAAA-CUUGGUGG--AAACACAGCCGCAACUUUA-GCGGCG--ACAUUUCCUCG-GGGCUUUGCCGCAAAUUAUGCGGUCGGGG-GCGUCUU ..(((.---(((((((((((....-)...(((.--...))).(((((.......-))))).--.))))..))))-)).((..((((((.....))))))..)))-))..... ( -43.20, z-score = -3.03, R) >droYak2.chr2R 18520726 101 - 21139217 CUGCCG---CUCGAAAAUGGGAAA-CUUCGAGG--AAACACAGCCGCAACUUUA-GCGGCG--ACAUUUCCUCG-GGGCUUUGCCGCAAAUUAUGCGGUCGGGG-ACGUCUU ..((((---(.......(((.(((-((((((((--(((....(((((.......-))))).--...))))))))-))).))).)))........)))))((...-.)).... ( -41.46, z-score = -3.26, R) >droSec1.super_1 4129656 101 + 14215200 CUGCCG---CUCGAGAAUGGGAAA-CUUGGUGG--AAACACAGCCGCAACUUUA-GCGGCG--ACAUUUCCUCA-GGGCUUUGCCGCAAAUUAUGCGGUCGGGG-ACGUCUU ..(((.---((.((((((((....-)...(((.--...))).(((((.......-))))).--.))))..))))-))))...((((((.....))))))((...-.)).... ( -37.80, z-score = -1.99, R) >droSim1.chr2R 5154196 101 + 19596830 CUGCCG---CUCGAGAAUGGGAAA-CUUGGUGG--AAACACAGCCGCAACUUUA-GCGGCG--ACAUUUCCUCA-GGGCUUUGCCGCAAAUUAUGCGGUCGGGG-ACGUCUU ..(((.---((.((((((((....-)...(((.--...))).(((((.......-))))).--.))))..))))-))))...((((((.....))))))((...-.)).... ( -37.80, z-score = -1.99, R) >droPer1.super_4 671473 107 + 7162766 CUGCCGGGUCUGCAGGAUAGGAAA-CUUUGAGG--AAACACAGCCGCAACUUUA-GCGGCGGGACAUUUCCUCU-GGGCUUUGCUGCAAAUUAUGCGGUGGGGCCAUGUCUU ((((.......))))(((((....-)...((((--(((..(.(((((.......-))))).)....))))))).-.(((((..(((((.....)))))..))))).)))).. ( -43.50, z-score = -2.77, R) >dp4.chr3 3685173 107 - 19779522 CUGCCGGGUCUGCAGGAUAGGAAA-CUUUGAGG--AAACACAGCCGCAACUUUA-GCGGCGGGACAUUUCCUCU-GGGCUUUGCUGCAAAUUAUGCGGUGGGGCCAUGUCUU ((((.......))))(((((....-)...((((--(((..(.(((((.......-))))).)....))))))).-.(((((..(((((.....)))))..))))).)))).. ( -43.50, z-score = -2.77, R) >droVir3.scaffold_12875 16519085 84 + 20611582 --AUUAUGCUUUGGGGCUGGAAAAACUUUGAAGC----------CGCAACUUUGCGCGGCG--ACAUUUCCUCUUGGGUCUUGACGCAAAUUAUGCGG-------------- --....(((.(..(((((.((.......((..((----------(((........))))).--.)).......)).)))))..).)))..........-------------- ( -24.94, z-score = -1.49, R) >droGri2.scaffold_15245 5364755 92 + 18325388 --AGUAUGCUCUAGGGCUAGAAAAACUUUGAAGCAACACAGAGCCGCAACUUUACGCGGCG--ACAUUUCCUCU-GGGCCUUGACGCAAAUUAUGCG--------------- --....(((((.(((.(((((.....((((........))))(((((........))))).--........)))-)).))).)).))).........--------------- ( -26.80, z-score = -1.64, R) >consensus CUGCCG___CUCGAGAAUGGGAAA_CUUUGAGG__AAACACAGCCGCAACUUUA_GCGGCG__ACAUUUCCUCU_GGGCUUUGCCGCAAAUUAUGCGGUCGGGG_ACGUCUU ..(((....(((......))).....................(((((........)))))................)))...((((((.....))))))............. (-17.51 = -17.42 + -0.09)
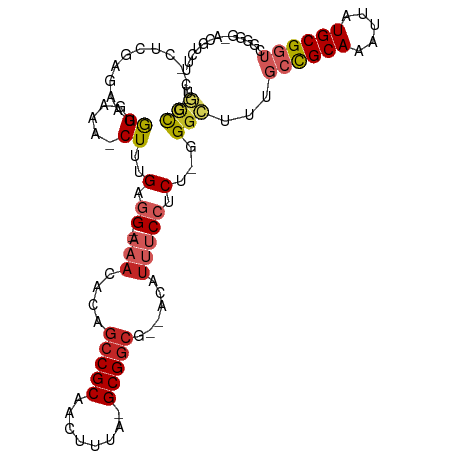
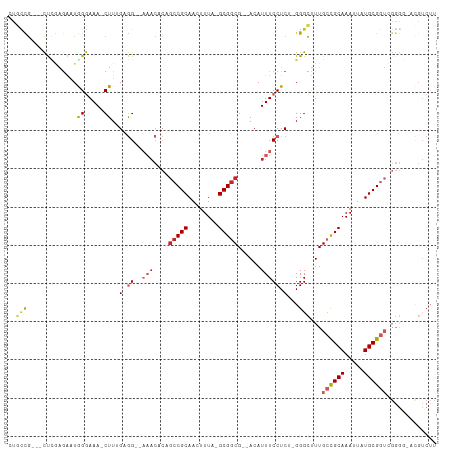
| Location | 6,542,307 – 6,542,409 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Shannon entropy | 0.46096 |
| G+C content | 0.53825 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.623972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6542307 102 - 21146708 AAGACGU-CCCCGACCGCAUAAUUUGCGGCAAAGCCC-UGAGGAAAUGU--CGCCGC-UAAAGUUGCGGCUGUGUUU-CCCACCAAG-UUUCCCAUUCUCGAG---CGGCAG .......-....(.(((((.....))))))...(((.-((.(((((..(--.(((((-.......))))).)..)))-))))....(-(((.........)))---)))).. ( -32.20, z-score = -1.76, R) >droAna3.scaffold_13266 18031824 101 - 19884421 AAGACAU-CCCCGACCGCAUAAUUUGCGGCAAUGCCC-GGAGGAAAUGU--CGCCGC-UAAAGUUGCGGCUGUGUUG--CCUCAAAG-UUUCCUAUCCUCGGGCUUCAA--- .......-....(.(((((.....))))))...((((-(((((.((..(--.(((((-.......))))).)..)).--))))..((-....)).....))))).....--- ( -35.60, z-score = -2.28, R) >droEre2.scaffold_4845 3362175 101 + 22589142 AAGACGC-CCCCGACCGCAUAAUUUGCGGCAAAGCCC-CGAGGAAAUGU--CGCCGC-UAAAGUUGCGGCUGUGUUU--CCACCAAG-UUUCCCAUUCUCGAG---CGGCAA .....((-((.((((((((.....)))))........-...(((((..(--.(((((-.......))))).)..)))--))......-..........))).)---.))).. ( -34.80, z-score = -2.38, R) >droYak2.chr2R 18520726 101 + 21139217 AAGACGU-CCCCGACCGCAUAAUUUGCGGCAAAGCCC-CGAGGAAAUGU--CGCCGC-UAAAGUUGCGGCUGUGUUU--CCUCGAAG-UUUCCCAUUUUCGAG---CGGCAG .....((-((.((((((((.....)))))..((((..-((((((((..(--.(((((-.......))))).)..)))--)))))..)-))).......))).)---.))).. ( -40.10, z-score = -3.92, R) >droSec1.super_1 4129656 101 - 14215200 AAGACGU-CCCCGACCGCAUAAUUUGCGGCAAAGCCC-UGAGGAAAUGU--CGCCGC-UAAAGUUGCGGCUGUGUUU--CCACCAAG-UUUCCCAUUCUCGAG---CGGCAG .......-....(.(((((.....))))))...(((.-...(((((..(--.(((((-.......))))).)..)))--)).....(-(((.........)))---)))).. ( -32.40, z-score = -1.79, R) >droSim1.chr2R 5154196 101 - 19596830 AAGACGU-CCCCGACCGCAUAAUUUGCGGCAAAGCCC-UGAGGAAAUGU--CGCCGC-UAAAGUUGCGGCUGUGUUU--CCACCAAG-UUUCCCAUUCUCGAG---CGGCAG .......-....(.(((((.....))))))...(((.-...(((((..(--.(((((-.......))))).)..)))--)).....(-(((.........)))---)))).. ( -32.40, z-score = -1.79, R) >droPer1.super_4 671473 107 - 7162766 AAGACAUGGCCCCACCGCAUAAUUUGCAGCAAAGCCC-AGAGGAAAUGUCCCGCCGC-UAAAGUUGCGGCUGUGUUU--CCUCAAAG-UUUCCUAUCCUGCAGACCCGGCAG .......((.....))((....(((((((..((((..-.(((((((..(...(((((-.......))))).)..)))--))))...)-)))......)))))))....)).. ( -31.90, z-score = -1.85, R) >dp4.chr3 3685173 107 + 19779522 AAGACAUGGCCCCACCGCAUAAUUUGCAGCAAAGCCC-AGAGGAAAUGUCCCGCCGC-UAAAGUUGCGGCUGUGUUU--CCUCAAAG-UUUCCUAUCCUGCAGACCCGGCAG .......((.....))((....(((((((..((((..-.(((((((..(...(((((-.......))))).)..)))--))))...)-)))......)))))))....)).. ( -31.90, z-score = -1.85, R) >droVir3.scaffold_12875 16519085 84 - 20611582 --------------CCGCAUAAUUUGCGUCAAGACCCAAGAGGAAAUGU--CGCCGCGCAAAGUUGCG----------GCUUCAAAGUUUUUCCAGCCCCAAAGCAUAAU-- --------------(((((...(((((((...(((((....))....))--)...)))))))..))))----------)................((......)).....-- ( -21.40, z-score = -1.77, R) >droGri2.scaffold_15245 5364755 92 - 18325388 ---------------CGCAUAAUUUGCGUCAAGGCCC-AGAGGAAAUGU--CGCCGCGUAAAGUUGCGGCUCUGUGUUGCUUCAAAGUUUUUCUAGCCCUAGAGCAUACU-- ---------------((((...(((((((...(((.(-(.......)).--.))))))))))..))))((((((....(((.............)))..)))))).....-- ( -24.42, z-score = -0.21, R) >consensus AAGACGU_CCCCGACCGCAUAAUUUGCGGCAAAGCCC_AGAGGAAAUGU__CGCCGC_UAAAGUUGCGGCUGUGUUU__CCUCAAAG_UUUCCCAUCCUCGAG___CGGCAG ..............(((((.....)))))............(((((......(((((........)))))...(........).....)))))................... (-13.38 = -13.92 + 0.54)

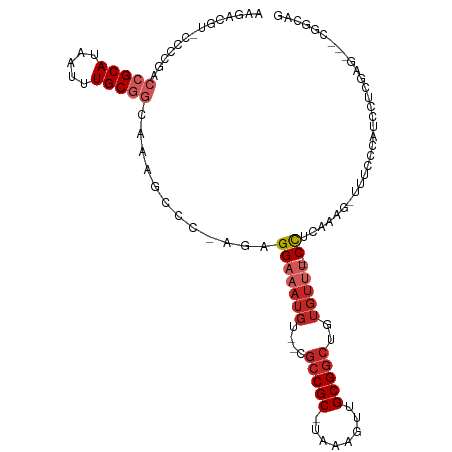
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:54 2011