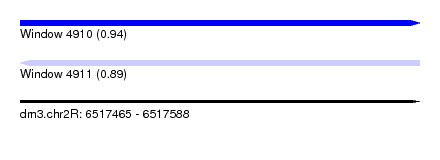
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,517,465 – 6,517,588 |
| Length | 123 |
| Max. P | 0.940439 |

| Location | 6,517,465 – 6,517,588 |
|---|---|
| Length | 123 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 56.85 |
| Shannon entropy | 0.59722 |
| G+C content | 0.49879 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -21.41 |
| Energy contribution | -19.53 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
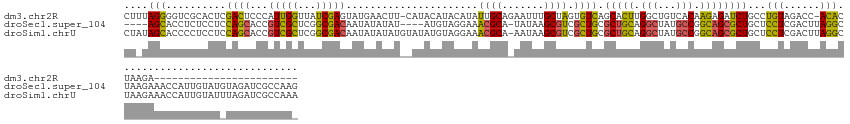
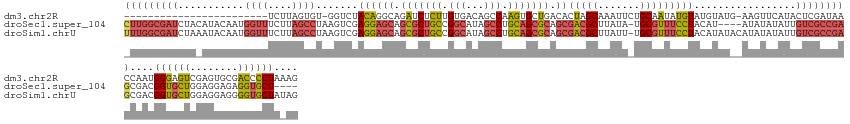
>dm3.chr2R 6517465 123 + 21146708 CUUUAGGGGUCGCACUCGACUCCCAUUGGUUAUCGAGUAUGAACUU-CAUACAUACAUAUUGCAGAAUUUGCUAGUGUCAGCACUUGGCUGUCACAAGAGAUCUGCCUGUAGACC-ACACUAAGA------------------------ .....(((((((....)))).)))..(((((.....(((((.....-))))).((((....(((((.(((.((.(((.((((.....)))).))).)))))))))).))))))))-)........------------------------ ( -39.40, z-score = -2.80, R) >droSec1.super_104 52315 140 - 95517 ----AGCACCUCUCCUCCAGCACCGUCGCUCGGCGACAAUAUAUAU----AUGUAGGAAACGCA-UAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAUGUAGAUCGCCAAG ----..............(((......))).(((((...(((((((----(((.((((.((((.-.....)))).((.(((((((.(((...))).))))))).)))))))).....((........))..))))))))..)))))... ( -47.40, z-score = -1.39, R) >droSim1.chrU 5095458 148 + 15797150 CUAUAGCACCCCUCCUCCAGCACCGUCGCUCGGCGACAAUAUAUAUGUAUAUGUAGGAAACGCA-AAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAUUUAGAUCGCCAAA ..................(((......))).((((((...(((((......((.((((.((((.-.....)))).((.(((((((.(((...))).))))))).)))))))).....((........))..)))))...).)))))... ( -42.70, z-score = -0.31, R) >consensus CU_UAGCACCCCUCCUCCAGCACCGUCGCUCGGCGACAAUAUAUAU__AUAUGUAGGAAACGCA_AAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAACCAUUGUAU_UAGAUCGCCAA_ ....(((..........((((...(((((...)))))......................((((.......)))).)))).(((((.(((...))).))))))))...(((......))).............................. (-21.41 = -19.53 + -1.88)
| Location | 6,517,465 – 6,517,588 |
|---|---|
| Length | 123 |
| Sequences | 3 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 56.85 |
| Shannon entropy | 0.59722 |
| G+C content | 0.49879 |
| Mean single sequence MFE | -47.20 |
| Consensus MFE | -23.74 |
| Energy contribution | -27.53 |
| Covariance contribution | 3.79 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
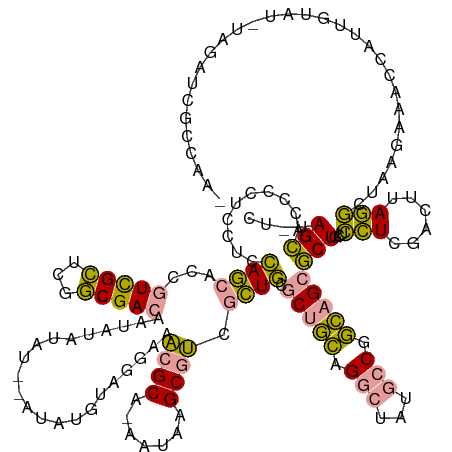
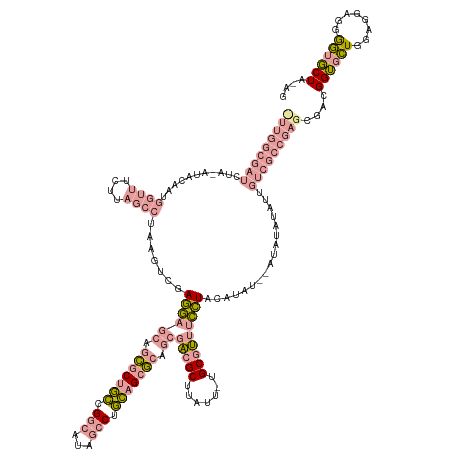
>dm3.chr2R 6517465 123 - 21146708 ------------------------UCUUAGUGU-GGUCUACAGGCAGAUCUCUUGUGACAGCCAAGUGCUGACACUAGCAAAUUCUGCAAUAUGUAUGUAUG-AAGUUCAUACUCGAUAACCAAUGGGAGUCGAGUGCGACCCCUAAAG ------------------------..((((.(.-((((((((.(((((...((.(((.((((.....)))).))).)).....)))))....))))......-.......((((((((..((...))..)))))))).))))))))).. ( -38.20, z-score = -2.31, R) >droSec1.super_104 52315 140 + 95517 CUUGGCGAUCUACAUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUA-UGCGUUUCCUACAU----AUAUAUAUUGUCGCCGAGCGACGGUGCUGGAGGAGAGGUGCU---- (((((((((...........((((....)))).......((((((.(((((((.(((...))).))))))).))(((((.....-.)))))))))....----.........)))))))))....((..((........))..))---- ( -51.90, z-score = -0.91, R) >droSim1.chrU 5095458 148 - 15797150 UUUGGCGAUCUAAAUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUU-UGCGUUUCCUACAUAUACAUAUAUAUUGUCGCCGAGCGACGGUGCUGGAGGAGGGGUGCUAUAG ..................(((((..(((..(((.(((..((((((.(((((((.(((...))).))))))).))(((((.....-.)))))))))..............((((((((...)))))))))))..)))..))).))))).. ( -51.50, z-score = -0.76, R) >consensus _UUGGCGAUCUA_AUACAAUGGUUUCUUAGCCUAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUU_UGCGUUUCCUACAUAU__AUAUAUAUUGUCGCCGAGCGACGGUGCUGGAGGAGGGGUGCUA_AG (((((((((...........(((......))).......((((((.(((((((.(((...))).))))))).))(((((.......))))))))).................)))))))))....((((((........)))))).... (-23.74 = -27.53 + 3.79)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:51 2011