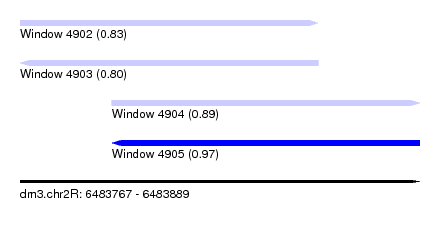
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,483,767 – 6,483,889 |
| Length | 122 |
| Max. P | 0.965250 |

| Location | 6,483,767 – 6,483,858 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Shannon entropy | 0.39072 |
| G+C content | 0.42985 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -15.29 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
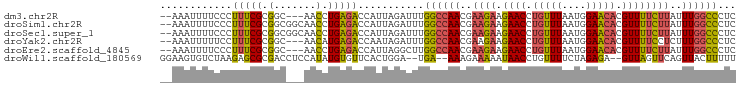
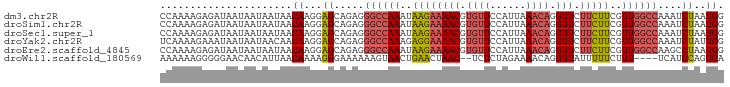
>dm3.chr2R 6483767 91 + 21146708 --AAAUUUUCCCUUUCGCGGC---AACCUGAGACCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUC --((((((....(((((.(..---..).))))).....))))))((((((..(((((((...(((((....)))))..)))))))..))))))... ( -23.80, z-score = -2.53, R) >droSim1.chr2R 5107812 94 + 19596830 --AAAUUUUCCCUUUCGCGGCGGCAACCUGAGACCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUC --((((((......((.(((.(....)))).)).....))))))((((((..(((((((...(((((....)))))..)))))))..))))))... ( -24.50, z-score = -1.73, R) >droSec1.super_1 4086169 94 + 14215200 --AAAUUUUCCCUUUCGCGGCGGCAACCUGAGACCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUC --((((((......((.(((.(....)))).)).....))))))((((((..(((((((...(((((....)))))..)))))))..))))))... ( -24.50, z-score = -1.73, R) >droYak2.chr2R 18475318 91 - 21139217 --AAAUUUUUCCUUUCGCGGC---AACAUGAGACCAAUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCCUCUUUGGCCCUC --((((((....(((((.(..---..).))))).....))))))((((((.((.(((((...(((((....)))))..))))).)).))))))... ( -23.40, z-score = -2.17, R) >droEre2.scaffold_4845 3315053 91 - 22589142 --AAAUUUUCCCUUUCGCGGC---AACCUGAGACCAUUAGGCUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUC --..........(((((.(..---..).)))))((....))...((((((..(((((((...(((((....)))))..)))))))..))))))... ( -22.60, z-score = -1.31, R) >droWil1.scaffold_180569 381475 90 + 1405142 GGAAGUGUCUAAGAGCGCGACCUCCAUAUGUGUUCACUGGA--UGA--AAAGAAAAAUAACCUGUUUUCUAGAGA--GUUAGUUCAGUUACUUUUU (((((((.((..((((..(((((((((..((....))...)--)).--..((((((........)))))).))).--))).)))))).))))))). ( -16.90, z-score = 0.09, R) >consensus __AAAUUUUCCCUUUCGCGGC___AACCUGAGACCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUC ............(((((.((......)))))))...........((((((..((((.((((.(((((....))))).))))))))..))))))... (-15.29 = -15.02 + -0.27)
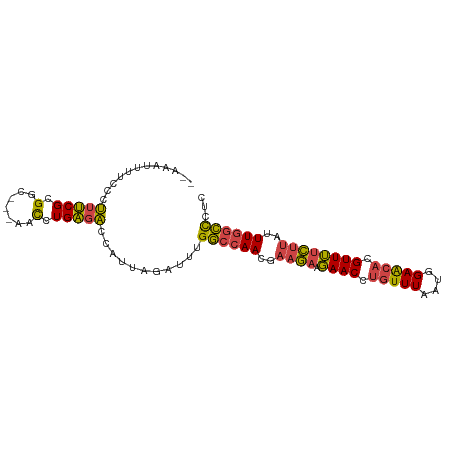
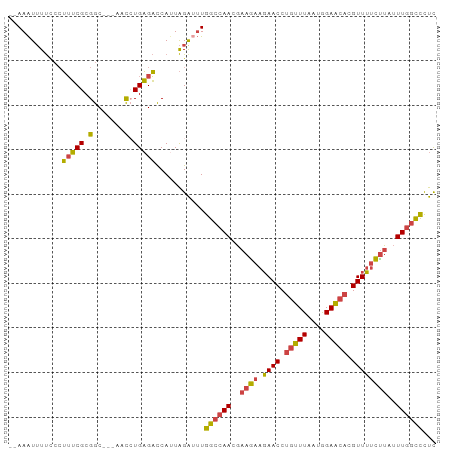
| Location | 6,483,767 – 6,483,858 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Shannon entropy | 0.39072 |
| G+C content | 0.42985 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.20 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6483767 91 - 21146708 GAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGGUCUCAGGUU---GCCGCGAAAGGGAAAAUUU-- (((((((((..((((((((.((((......)))).))).)))))..))))))..(((....)))))).....---.((.(....))).......-- ( -25.90, z-score = -2.10, R) >droSim1.chr2R 5107812 94 - 19596830 GAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGGUCUCAGGUUGCCGCCGCGAAAGGGAAAAUUU-- ((.((((((..((((((((.((((......)))).))).)))))..))))))...)).(((..((((...((((....))))..))))..))).-- ( -27.50, z-score = -1.85, R) >droSec1.super_1 4086169 94 - 14215200 GAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGGUCUCAGGUUGCCGCCGCGAAAGGGAAAAUUU-- ((.((((((..((((((((.((((......)))).))).)))))..))))))...)).(((..((((...((((....))))..))))..))).-- ( -27.50, z-score = -1.85, R) >droYak2.chr2R 18475318 91 + 21139217 GAGGGCCAAAGAGGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAUUGGUCUCAUGUU---GCCGCGAAAGGAAAAAUUU-- ((.((((((.(((((((((.((((......)))).))).)))))).))))))...))....(((........---))).(....).........-- ( -26.90, z-score = -2.21, R) >droEre2.scaffold_4845 3315053 91 + 22589142 GAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAGCCUAAUGGUCUCAGGUU---GCCGCGAAAGGGAAAAUUU-- (((((((((..((((((((.((((......)))).))).)))))..))))))..(((....)))))).....---.((.(....))).......-- ( -28.80, z-score = -2.35, R) >droWil1.scaffold_180569 381475 90 - 1405142 AAAAAGUAACUGAACUAAC--UCUCUAGAAAACAGGUUAUUUUUCUUU--UCA--UCCAGUGAACACAUAUGGAGGUCGCGCUCUUAGACACUUCC .((((((((((...(((..--....)))......))))))))))...(--(((--(...))))).......(((((((.........))).)))). ( -12.50, z-score = 0.73, R) >consensus GAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGGUCUCAGGUU___GCCGCGAAAGGGAAAAUUU__ (((((((((..((((((((.((((......)))).))).)))))..))))))..(((....)))))).........((.(....)))......... (-16.14 = -17.20 + 1.06)

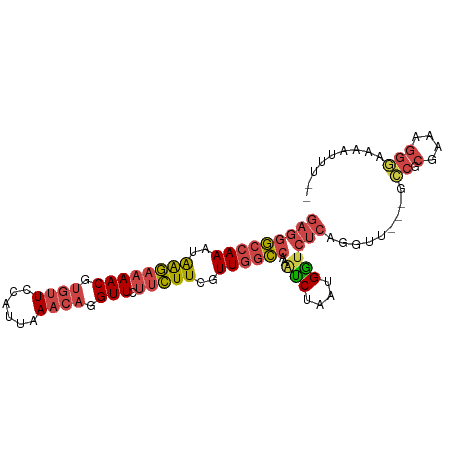
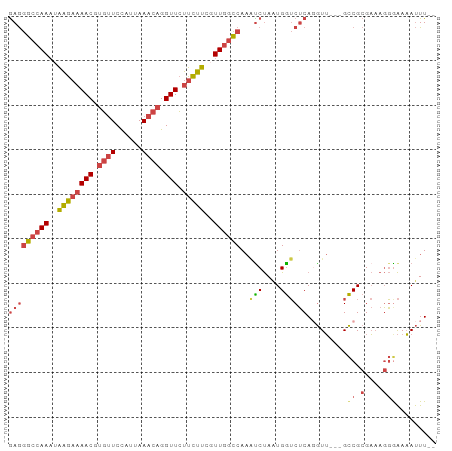
| Location | 6,483,795 – 6,483,889 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Shannon entropy | 0.34473 |
| G+C content | 0.36863 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6483795 94 + 21146708 CCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUCUGCUCCUUGUUAUUAUUAUUAUCUCUUUUGG (((..(((...((((((..(((((((...(((((....)))))..)))))))..))))))....((.....)).............)))..))) ( -21.90, z-score = -2.69, R) >droSim1.chr2R 5107843 94 + 19596830 CCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUCUGCUCCUUGUUAUUAUUAUUAUCUCUUUUGG (((..(((...((((((..(((((((...(((((....)))))..)))))))..))))))....((.....)).............)))..))) ( -21.90, z-score = -2.69, R) >droSec1.super_1 4086200 94 + 14215200 CCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUCUGCUCCUUGUUAUUAUUAUUAUCUCUUUUGG (((..(((...((((((..(((((((...(((((....)))))..)))))))..))))))....((.....)).............)))..))) ( -21.90, z-score = -2.69, R) >droYak2.chr2R 18475346 94 - 21139217 CCAAUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCCUCUUUGGCCCUCUGCUCCUUGUUGUUAUUAUUAUUUCUUUUGA .(((.(((...((((((.((.(((((...(((((....)))))..))))).)).))))))....((.....)).............))).))). ( -19.90, z-score = -1.87, R) >droEre2.scaffold_4845 3315081 94 - 22589142 CCAUUAGGCUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUCUGCUCCUUGUUAUUAUUAUUAUCUCUUUUGG .((...(((..((((((..(((((((...(((((....)))))..)))))))..))))))....)))...))...................... ( -21.20, z-score = -1.79, R) >droWil1.scaffold_180569 381508 88 + 1405142 UCACUGGAUGA----AAAGAAAAAUAACCUGUUUUCUAGAGA--GUUAGUUCAGUUACUUUUUUCCCUUUUGUUAAUGUUGUUCCCCCUUUUUU ..((..((..(----((.((((((....(((....((((...--.))))..))).....))))))..)))..))...))............... ( -8.40, z-score = 1.39, R) >consensus CCAUUAGAUUUGGCCAACGAAGAAGAACCUGUUUAAUGGAACACGUUUUCUUAUUUGGCCCUCUGCUCCUUGUUAUUAUUAUUAUCUCUUUUGG ....((((...((((((..((((.((((.(((((....))))).))))))))..)))))).))))............................. (-14.21 = -14.77 + 0.56)
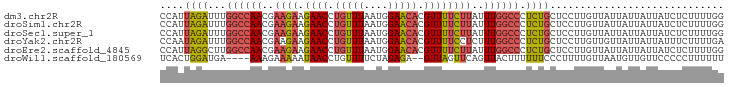

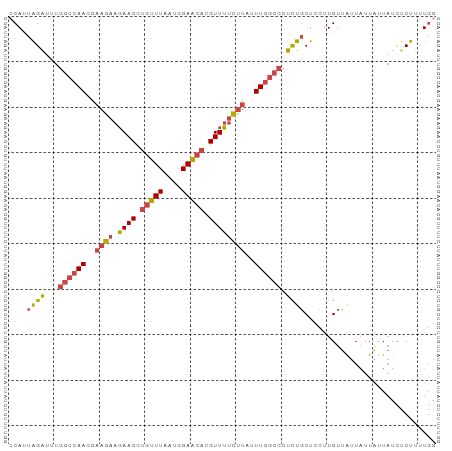
| Location | 6,483,795 – 6,483,889 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Shannon entropy | 0.34473 |
| G+C content | 0.36863 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6483795 94 - 21146708 CCAAAAGAGAUAAUAAUAAUAACAAGGAGCAGAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGG ..............................(((.((((((..((((((((.((((......)))).))).)))))..))))))...)))..... ( -22.10, z-score = -2.63, R) >droSim1.chr2R 5107843 94 - 19596830 CCAAAAGAGAUAAUAAUAAUAACAAGGAGCAGAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGG ..............................(((.((((((..((((((((.((((......)))).))).)))))..))))))...)))..... ( -22.10, z-score = -2.63, R) >droSec1.super_1 4086200 94 - 14215200 CCAAAAGAGAUAAUAAUAAUAACAAGGAGCAGAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGG ..............................(((.((((((..((((((((.((((......)))).))).)))))..))))))...)))..... ( -22.10, z-score = -2.63, R) >droYak2.chr2R 18475346 94 + 21139217 UCAAAAGAAAUAAUAAUAACAACAAGGAGCAGAGGGCCAAAGAGGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAUUGG ..............................(((.((((((.(((((((((.((((......)))).))).)))))).))))))...)))..... ( -25.30, z-score = -3.56, R) >droEre2.scaffold_4845 3315081 94 + 22589142 CCAAAAGAGAUAAUAAUAAUAACAAGGAGCAGAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAGCCUAAUGG .........................((.((....((((((..((((((((.((((......)))).))).)))))..))))))..))))..... ( -21.90, z-score = -2.05, R) >droWil1.scaffold_180569 381508 88 - 1405142 AAAAAAGGGGGAACAACAUUAACAAAAGGGAAAAAAGUAACUGAACUAAC--UCUCUAGAAAACAGGUUAUUUUUCUUU----UCAUCCAGUGA ...(((((..(((.(((.........(((((....(((......)))...--))))).........))).)))..))))----).......... ( -12.17, z-score = 0.51, R) >consensus CCAAAAGAGAUAAUAAUAAUAACAAGGAGCAGAGGGCCAAAUAAGAAAACGUGUUCCAUUAAACAGGUUCUUCUUCGUUGGCCAAAUCUAAUGG ......................((...((.....((((((..((((((((.((((......)))).))).)))))..))))))....))..)). (-15.10 = -15.77 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:45 2011