
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,462,662 – 6,462,821 |
| Length | 159 |
| Max. P | 0.997557 |

| Location | 6,462,662 – 6,462,754 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 85.44 |
| Shannon entropy | 0.26300 |
| G+C content | 0.38873 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
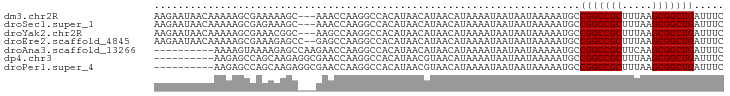
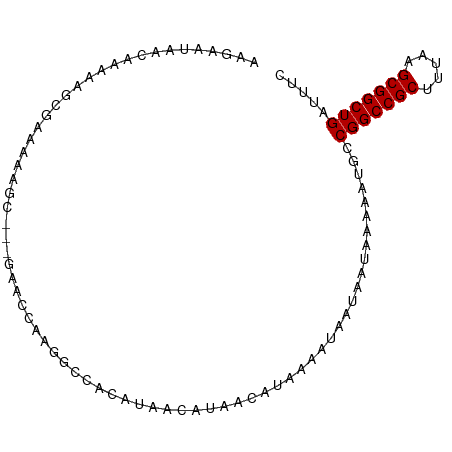
>dm3.chr2R 6462662 92 + 21146708 AAGAAUAACAAAAAGCGAAAAAGC---AAACCAAGGCCACAUAACAUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC ..............((......))---............................................(((((((.....)))))))..... ( -15.60, z-score = -1.59, R) >droSec1.super_1 4073560 92 + 14215200 AAGAAUAACAAAAAGCGAGAAAGC---AAACCAAGGCCACAUAACAUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC ..............((......))---............................................(((((((.....)))))))..... ( -15.60, z-score = -1.51, R) >droYak2.chr2R 18462508 92 - 21139217 AAGAAUAACAAAAAGCGAAACGGC---AAGCCAAGGCCACAUAACAUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC ................((((.(((---........))).................................(((((((.....))))))).)))) ( -18.20, z-score = -1.54, R) >droEre2.scaffold_4845 3302001 93 - 22589142 AAGAAUAACAAAAAGCGAAAGAGCC--GAGCCAAGGCCACAUAACAUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC ...............(....).(((--.......)))..................................(((((((.....)))))))..... ( -18.40, z-score = -1.68, R) >droAna3.scaffold_13266 17959933 85 + 19884421 ----------AAAAGUAAAAGAGCCAAGAACCAAGGCCACAUAACAUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUCAAGCGGCUGAUUUC ----------..........(.(((.........)))).................................(((((((.....)))))))..... ( -16.70, z-score = -2.18, R) >dp4.chr3 3633625 85 - 19779522 ----------AAGAGCCAGCAAGAGGCGAACCAAGGCCACAUAACGUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC ----------........(((...(((........)))((.....)).....................)))(((((((.....)))))))..... ( -19.20, z-score = -1.21, R) >droPer1.super_4 619035 85 + 7162766 ----------AAGAGCCAGCAAGAGGCGAACCAAGGCCACAUAACGUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC ----------........(((...(((........)))((.....)).....................)))(((((((.....)))))))..... ( -19.20, z-score = -1.21, R) >consensus AAGAAUAACAAAAAGCGAAAAAGC___GAACCAAGGCCACAUAACAUAACAUAAAAUAAUAAUAAAAAUGCCGGCCGCUUUAAGCGGCUGAUUUC .......................................................................(((((((.....)))))))..... (-13.50 = -13.50 + 0.00)
| Location | 6,462,722 – 6,462,821 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 63.32 |
| Shannon entropy | 0.66318 |
| G+C content | 0.49399 |
| Mean single sequence MFE | -16.41 |
| Consensus MFE | -9.03 |
| Energy contribution | -9.31 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
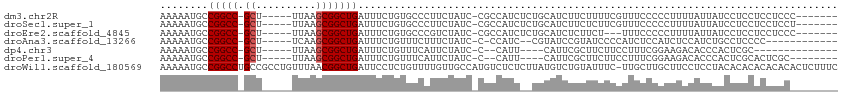
>dm3.chr2R 6462722 99 + 21146708 AAAAAUGCCGGCC-GCU-----UUAAGCGGCUGAUUUCUGUGCCCUUCUAUC-CGCCAUCUCUGCAUCUUCUUUUCGUUUCCCCCUUUUAUUAUCCUCCUCCUCCC------- ....(((((((((-((.-----....))))))).....((.((.........-.)))).....)))).......................................------- ( -15.80, z-score = -2.56, R) >droSec1.super_1 4073620 99 + 14215200 AAAAAUGCCGGCC-GCU-----UUAAGCGGCUGAUUUCUGUGCCCUUCUAUC-CGCCAUCUCUGCAUCUUCUCUUCGUUUCCCCCUUUUAUUAUCCUCCUCCUCCU------- ....(((((((((-((.-----....))))))).....((.((.........-.)))).....)))).......................................------- ( -15.80, z-score = -2.58, R) >droEre2.scaffold_4845 3302062 96 - 22589142 AAAAAUGCCGGCC-GCU-----UUAAGCGGCUGAUUUCUGUGCCCGUCUAUC-CGCCAUCUCUGCAUCUCUUCU---UUUCCCCCUUUUAUUAUCCUCCUCCUCCC------- ....(((((((((-((.-----....))))))).....((.((.........-.)))).....)))).......---.............................------- ( -15.80, z-score = -2.57, R) >droAna3.scaffold_13266 17959986 91 + 19884421 AAAAAUGCCGGCC-GCU-----UCAAGCGGCUGAUUUCUGUUUCUUUCUAUC-C-CCAUC--CGUAUCCGUAUCCCCAUCUCCAUCUCCAUCUGCCUCCCC------------ ........(((((-((.-----....)))))))...................-.-.....--.......................................------------ ( -13.50, z-score = -2.21, R) >dp4.chr3 3633678 86 - 19779522 AAAAAUGCCGGCC-GCU-----UUAAGCGGCUGAUUUCUGUUUCAUUCUAUC-C--CAUU----CAUUCGCUUCUUCCUUUCGGAAGACACCCACUCGC-------------- ........(((((-((.-----....)))))))...................-.--....----........((((((....))))))...........-------------- ( -20.60, z-score = -2.46, R) >droPer1.super_4 619088 92 + 7162766 AAAAAUGCCGGCC-GCU-----UUAAGCGGCUGAUUUCUGUUUCAUUCUAUC-C--CAUU----CAUUCGCUUCUUCCUUUCGGAAGACACCCACUCGCACUCGC-------- .....((((((((-((.-----....)))))))...................-.--....----........((((((....)))))).........))).....-------- ( -22.90, z-score = -2.77, R) >droWil1.scaffold_180569 365069 112 + 1405142 AAAAAUGCCGGCCUGCCGCCUGUUUAACGGCUGAUUCCUCUGUUUUGUUGCCAUGUCUCUCUUAUGUCUGUAUUUC-UUGCUUGCUUCCUCCUACACACACACACACUCUUUC ......((.(((..((((.........))))....................((((.......))))..........-..))).))............................ ( -10.50, z-score = 1.56, R) >consensus AAAAAUGCCGGCC_GCU_____UUAAGCGGCUGAUUUCUGUGUCCUUCUAUC_CGCCAUC__UGCAUCUGCUUUUCCUUUCCCCCUUUCAUCAACCUCCUCCUCC________ ........(((((.(((........))))))))................................................................................ ( -9.03 = -9.31 + 0.29)
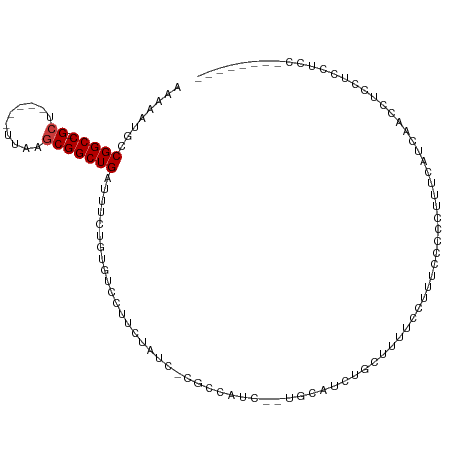
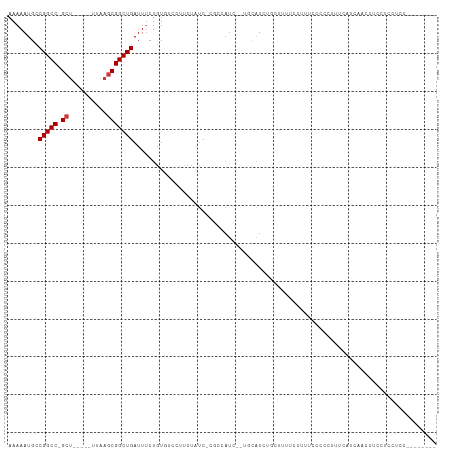
| Location | 6,462,722 – 6,462,821 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 63.32 |
| Shannon entropy | 0.66318 |
| G+C content | 0.49399 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -6.81 |
| Energy contribution | -6.44 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.515571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6462722 99 - 21146708 -------GGGAGGAGGAGGAUAAUAAAAGGGGGAAACGAAAAGAAGAUGCAGAGAUGGCG-GAUAGAAGGGCACAGAAAUCAGCCGCUUAA-----AGC-GGCCGGCAUUUUU -------........................(....).....((((((((.((..((((.-.........)).))....)).(((((....-----.))-)))..)))))))) ( -20.90, z-score = -2.06, R) >droSec1.super_1 4073620 99 - 14215200 -------AGGAGGAGGAGGAUAAUAAAAGGGGGAAACGAAGAGAAGAUGCAGAGAUGGCG-GAUAGAAGGGCACAGAAAUCAGCCGCUUAA-----AGC-GGCCGGCAUUUUU -------........................(....).....((((((((.((..((((.-.........)).))....)).(((((....-----.))-)))..)))))))) ( -20.90, z-score = -2.03, R) >droEre2.scaffold_4845 3302062 96 + 22589142 -------GGGAGGAGGAGGAUAAUAAAAGGGGGAAA---AGAAGAGAUGCAGAGAUGGCG-GAUAGACGGGCACAGAAAUCAGCCGCUUAA-----AGC-GGCCGGCAUUUUU -------.............................---...((((((((.((..((((.-.........)).))....)).(((((....-----.))-)))..)))))))) ( -19.10, z-score = -1.14, R) >droAna3.scaffold_13266 17959986 91 - 19884421 ------------GGGGAGGCAGAUGGAGAUGGAGAUGGGGAUACGGAUACG--GAUGG-G-GAUAGAAAGAAACAGAAAUCAGCCGCUUGA-----AGC-GGCCGGCAUUUUU ------------..................(((((((......((....))--.....-.-..................((.(((((....-----.))-))).))))))))) ( -14.50, z-score = 0.48, R) >dp4.chr3 3633678 86 + 19779522 --------------GCGAGUGGGUGUCUUCCGAAAGGAAGAAGCGAAUG----AAUG--G-GAUAGAAUGAAACAGAAAUCAGCCGCUUAA-----AGC-GGCCGGCAUUUUU --------------..(((((.((.((((((....)))))).)).....----....--.-..................((.(((((....-----.))-))).))))))).. ( -25.40, z-score = -1.82, R) >droPer1.super_4 619088 92 - 7162766 --------GCGAGUGCGAGUGGGUGUCUUCCGAAAGGAAGAAGCGAAUG----AAUG--G-GAUAGAAUGAAACAGAAAUCAGCCGCUUAA-----AGC-GGCCGGCAUUUUU --------..((((((......((.((((((....)))))).)).....----....--.-.....................(((((....-----.))-)))..)))))).. ( -28.60, z-score = -2.04, R) >droWil1.scaffold_180569 365069 112 - 1405142 GAAAGAGUGUGUGUGUGUGUAGGAGGAAGCAAGCAA-GAAAUACAGACAUAAGAGAGACAUGGCAACAAAACAGAGGAAUCAGCCGUUAAACAGGCGGCAGGCCGGCAUUUUU ..((((((((.(((((.((((.......(....)..-....)))).))))).........((((.........((....)).((((((.....))))))..)))))))))))) ( -26.82, z-score = -2.30, R) >consensus ________GGAGGAGGAGGAUGAUGAAAGCGGGAAAGGAAAAGCAGAUGCA__GAUGGCG_GAUAGAAGGACACAGAAAUCAGCCGCUUAA_____AGC_GGCCGGCAUUUUU ..........................................................................(((((((.((((((........))).)))..).)))))) ( -6.81 = -6.44 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:41 2011