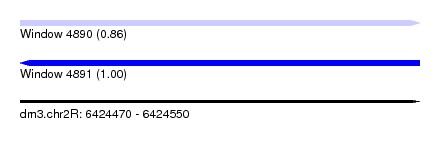
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,424,470 – 6,424,550 |
| Length | 80 |
| Max. P | 0.995058 |

| Location | 6,424,470 – 6,424,550 |
|---|---|
| Length | 80 |
| Sequences | 11 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 71.50 |
| Shannon entropy | 0.58010 |
| G+C content | 0.32575 |
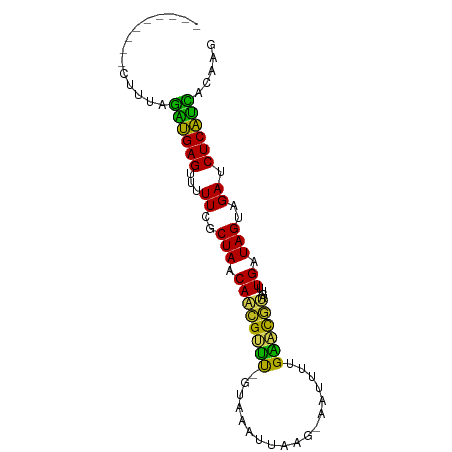
| Mean single sequence MFE | -11.37 |
| Consensus MFE | -8.86 |
| Energy contribution | -7.46 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6424470 80 + 21146708 CUUGUGAUGAGAUCUACUAUCAAACUACGUUCAAAAUUUGUUACAUUACGAAACGUUGUUAGCGAAAAACUCAUCUAAUA--------- .....((((((.........................(((((......))))).(((.....))).....)))))).....--------- ( -11.60, z-score = -0.58, R) >droSim1.chr2R 5055529 79 + 19596830 CUUGUGAUGAGAUCUACUAUCAAACUACGUUCAAAAUU-UUCACUUUACGAAACGUUGUUAGCGAAAAACUCAUCUAAAU--------- .....((((((...........(((...))).....((-(((.((..((((....)))).)).))))).)))))).....--------- ( -10.70, z-score = -0.72, R) >droSec1.super_1 4035965 79 + 14215200 CUUGUGUUGAGAUCUACUAUCAAACUACGUUCAAAAUU-CUCAUUUUACGAAACGUUGUUAGCGAAAAACUCAACUUGGG--------- ((...((((((................(((..(((((.-...))))))))...(((.....))).....))))))..)).--------- ( -10.70, z-score = 0.07, R) >droYak2.chr2R 18422878 78 - 21139217 CUUGUGAUGAGAUCUACUAUCAAACUACGUUCAAAAU--GUUAAUUUAAGAAACGUUGUUAGCGAAAAACUCAUCUAAAU--------- .....((((((((.....)))......((((...(((--(((.........))))))...))))......))))).....--------- ( -10.50, z-score = -0.54, R) >droEre2.scaffold_4845 3261850 79 - 22589142 CUUGUGAUGAGAUCUACUAUCAAACUACGUUCAAAAUU-UUCAUUUUACGAAACGUUGUUAGCGAAAAACUCAUCUAAAU--------- .....((((((................(((..(((((.-...))))))))...(((.....))).....)))))).....--------- ( -10.50, z-score = -0.54, R) >droAna3.scaffold_13266 17899891 78 + 19884421 AUUGUAUUGAGAUCUACUAUCAAACUACGUUCAAAAUU-AUUAAAAUAC-GAACGUUGUUAGCGAAAAACUCAACUGGAA--------- ......(((((...........(((.((((((......-..........-)))))).))).........)))))......--------- ( -9.44, z-score = -0.12, R) >dp4.chr3 3599420 77 - 19779522 ACUGUGUUGAGAUCUACUAUCAAACUACGUUCAAUUUU-CAUUAUACG--AAACGUUGUUAGCGAAAAACUCAACUGAAG--------- .((..((((((................(((..(((...-.)))..)))--...(((.....))).....))))))...))--------- ( -10.40, z-score = 0.14, R) >droPer1.super_4 584682 77 + 7162766 ACUGUGUUGAGAUCUACUAUCAAACUACGUUCAAAUUU-CAUUUUACG--AAACGUUGUUAGCGAAAAACUCAACUGAAG--------- .((..((((((................(((........-......)))--...(((.....))).....))))))...))--------- ( -9.74, z-score = 0.47, R) >droWil1.scaffold_180569 328529 79 + 1405142 ACUGUAAAGAGAUCUACUAUCAAACUUUGCACUUUGUUUCAUUCUUUUC-AUGCAAUGUUAGCGAAAAACUCUUUUUUUG--------- ..(((((((.(((.....)))...))))))).((((((.(((((.....-..).))))..))))))..............--------- ( -8.70, z-score = 0.15, R) >droVir3.scaffold_12875 16411154 82 + 20611582 AUUGUGGAGAGAUCUACUAUCAAACUAAA-GCGUUAUCAAAAUGU---C-ACGCAAUGUUAGCGAAAAACUCUCCCAU--AUCAUGAUA ..((.((((((..................-(((((.....)))))---.-.(((.......))).....)))))))).--......... ( -15.20, z-score = -1.52, R) >droGri2.scaffold_15245 5259843 83 + 18325388 CUUGUGGAGAGAUCUACUAUCAAAC-GUA-GCGCUAUUCAAUUAU---C-ACGCAAUGUUAGCGAAAAACUCUCCCAUCAAAAAUGAUA ...(.((((((..((((........-)))-)(((((.........---.-.........))))).....)))))))((((....)))). ( -17.61, z-score = -2.33, R) >consensus CUUGUGAUGAGAUCUACUAUCAAACUACGUUCAAAAUU_CUUAAUUUAC_AAACGUUGUUAGCGAAAAACUCAUCUAAAG_________ .....((((((...........(((.(((((....................))))).))).........)))))).............. ( -8.86 = -7.46 + -1.40)

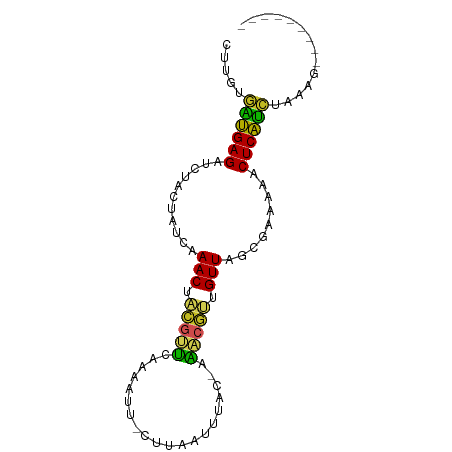
| Location | 6,424,470 – 6,424,550 |
|---|---|
| Length | 80 |
| Sequences | 11 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 71.50 |
| Shannon entropy | 0.58010 |
| G+C content | 0.32575 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -15.38 |
| Energy contribution | -13.40 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6424470 80 - 21146708 ---------UAUUAGAUGAGUUUUUCGCUAACAACGUUUCGUAAUGUAACAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAUCACAAG ---------.....((((((...((..(((...(((((....)))))..((((((........)))))).)))..)).))))))..... ( -16.50, z-score = -1.69, R) >droSim1.chr2R 5055529 79 - 19596830 ---------AUUUAGAUGAGUUUUUCGCUAACAACGUUUCGUAAAGUGAA-AAUUUUGAACGUAGUUUGAUAGUAGAUCUCAUCACAAG ---------.....((((((...((..(((((.((((((...(((((...-.))))))))))).))....)))..)).))))))..... ( -18.60, z-score = -2.29, R) >droSec1.super_1 4035965 79 - 14215200 ---------CCCAAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAAUGAG-AAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAAG ---------.....((((((...((..(((((.((((((...(((((...-.))))))))))).))....)))..)).))))))..... ( -16.70, z-score = -1.90, R) >droYak2.chr2R 18422878 78 + 21139217 ---------AUUUAGAUGAGUUUUUCGCUAACAACGUUUCUUAAAUUAAC--AUUUUGAACGUAGUUUGAUAGUAGAUCUCAUCACAAG ---------.....((((((...((..(((((.((((((...((((....--)))).)))))).))....)))..)).))))))..... ( -15.70, z-score = -2.15, R) >droEre2.scaffold_4845 3261850 79 + 22589142 ---------AUUUAGAUGAGUUUUUCGCUAACAACGUUUCGUAAAAUGAA-AAUUUUGAACGUAGUUUGAUAGUAGAUCUCAUCACAAG ---------.....((((((...((..(((((.((((((...(((((...-.))))))))))).))....)))..)).))))))..... ( -18.60, z-score = -2.51, R) >droAna3.scaffold_13266 17899891 78 - 19884421 ---------UUCCAGUUGAGUUUUUCGCUAACAACGUUC-GUAUUUUAAU-AAUUUUGAACGUAGUUUGAUAGUAGAUCUCAAUACAAU ---------.....((((((...((..(((((.((((((-(.(((.....-)))..))))))).))....)))..)).))))))..... ( -16.10, z-score = -2.41, R) >dp4.chr3 3599420 77 + 19779522 ---------CUUCAGUUGAGUUUUUCGCUAACAACGUUU--CGUAUAAUG-AAAAUUGAACGUAGUUUGAUAGUAGAUCUCAACACAGU ---------.....((((((...((..(((.((((....--(((.((((.-...)))).)))..).))).)))..)).))))))..... ( -18.10, z-score = -2.02, R) >droPer1.super_4 584682 77 - 7162766 ---------CUUCAGUUGAGUUUUUCGCUAACAACGUUU--CGUAAAAUG-AAAUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAGU ---------.....((((((...((..(((.....((((--(((...)))-))))(..(((...)))..))))..)).))))))..... ( -14.50, z-score = -0.60, R) >droWil1.scaffold_180569 328529 79 - 1405142 ---------CAAAAAAAGAGUUUUUCGCUAACAUUGCAU-GAAAAGAAUGAAACAAAGUGCAAAGUUUGAUAGUAGAUCUCUUUACAGU ---------.....((((((...((..(((((.((((((-.................)))))).))....)))..)).))))))..... ( -13.23, z-score = -0.74, R) >droVir3.scaffold_12875 16411154 82 - 20611582 UAUCAUGAU--AUGGGAGAGUUUUUCGCUAACAUUGCGU-G---ACAUUUUGAUAACGC-UUUAGUUUGAUAGUAGAUCUCUCCACAAU .........--.((((((((...((..(((.((..((((-.---...........))))-.......)).)))..)).)))))).)).. ( -18.20, z-score = -1.41, R) >droGri2.scaffold_15245 5259843 83 - 18325388 UAUCAUUUUUGAUGGGAGAGUUUUUCGCUAACAUUGCGU-G---AUAAUUGAAUAGCGC-UAC-GUUUGAUAGUAGAUCUCUCCACAAG (((((....)))))((((((...((..(((.((..((((-.---((......)).))))-...-...)).)))..)).))))))..... ( -20.80, z-score = -1.90, R) >consensus _________CUUUAGAUGAGUUUUUCGCUAACAACGUUU_GUAAAUUAAG_AAUUUUGAACGUAGUUUGAUAGUAGAUCUCAUCACAAG ..............((((((...((..(((((.((((((..................)))))).))....)))..)).))))))..... (-15.38 = -13.40 + -1.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:34 2011