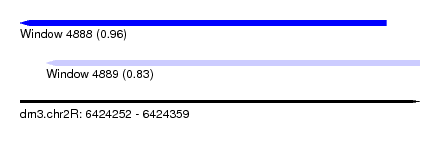
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,424,252 – 6,424,359 |
| Length | 107 |
| Max. P | 0.961557 |

| Location | 6,424,252 – 6,424,350 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.16 |
| Shannon entropy | 0.63484 |
| G+C content | 0.33405 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -15.06 |
| Energy contribution | -13.36 |
| Covariance contribution | -1.70 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
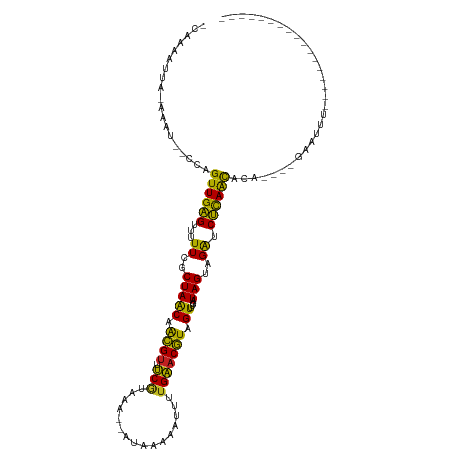
>dm3.chr2R 6424252 98 - 21146708 -CUAAACCA-AAGU--CCUGUUGAGUUUUUCGCUAACAACGUUUCGUAAA--GUGCAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACA----GAAUUUCUCGU------------ -........-....--..(((((((...((..(((((.((((((...(((--((....))))))))))).))....)))..)).)))))))..----...........------------ ( -17.00, z-score = -0.03, R) >droSim1.chr2R 5055335 99 - 19596830 -CUAAAAUA-AAAU--UCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAA--AUGAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACA----GAAUUACUCAUA----------- -........-.(((--((.((((((...((..(((((.((((((...(((--((....))))))))))).))....)))..)).))))))...----))))).......----------- ( -19.40, z-score = -1.64, R) >droSec1.super_1 4035788 93 - 14215200 ------------AU--UCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAA--AUAAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAGAAUGAAUUACUCAUG----------- ------------..--...((((((...((..(((((.((((((...(((--((....))))))))))).))....)))..)).)))))).....((((.....)))).----------- ( -16.80, z-score = -0.69, R) >droYak2.chr2R 18422682 88 + 21139217 -CUAAACCA-AAAU--CCAGUUGAGUUUUUCGCUAACAACGUUUCGU-AA--AUAUA-AAUUUGAACGUAGUUUGAUAGUAGAUCUUAACACA----GCA-------------------- -........-....--...((((((...((..(((((.((((((...-((--((...-.)))))))))).))....)))..)).))))))...----...-------------------- ( -12.70, z-score = -0.51, R) >droEre2.scaffold_4845 3261645 98 + 22589142 -CCAAACCA-AAAU--CCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAA--AUAUAUAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACA----GAAAUUCUGAU------------ -........-....--...((((((...((..(((((.((((((...(((--((....))))))))))).))....)))..)).)))))).((----(.....)))..------------ ( -18.10, z-score = -2.03, R) >droAna3.scaffold_13266 17899487 112 - 19884421 CUAAAAUUA-AACU--UCAGUUGAGUUUUUCGCUAACAACGUUUCUAAAA--ACAAUAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAAUACAA---GAUGUUAUAAGAGGGAUACUUAC .....((((-((((--...(((.(((.....)))))).((((((..(((.--.......))).))))))))))))))((((..((((...((..---...)).....))))..))))... ( -16.20, z-score = 0.31, R) >dp4.chr3 3599227 96 + 19779522 -CAAGAUUAUAUCU--UCAGUUGAGUUUUUCGCUAGCAACGUUUCGUAUA--AUG--AAAUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAA---GAGACCGAC-------------- -..........(((--(..((((((...((..(((..(((....(((.((--(..--....))).)))..)))...)))..)).))))))..))---)).......-------------- ( -17.10, z-score = 0.08, R) >droPer1.super_4 584489 96 - 7162766 -CAAGAUUAUAUCU--UCAGUUGAGUUUUUCGCUAACAACGUUUCGUAUA--AUG--AAAAUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAA---GAGACCGAC-------------- -..........(((--(..((((((...((..(((.((((....(((.((--((.--...)))).)))..).))).)))..)).))))))..))---)).......-------------- ( -19.60, z-score = -1.05, R) >droWil1.scaffold_180569 328358 92 - 1405142 --UAGAUUAAACCCAAAAAGUUGAGAUUUUCGCUAACAGUGUUCCA---------ACGCUUUUGGACAUGGUUUGAUAGUAGACCUCAACACAUUCUUCCGAU----------------- --..........((((((.((((.((....(((.....))).))))---------))..))))))....((((((....))))))..................----------------- ( -16.90, z-score = -0.72, R) >droVir3.scaffold_12875 16410578 98 - 20611582 CUAAAUUAAUAUCU--CAGGUUGGGUUUUUCGCUAACAACGUACCGCAUA--UGAA-AAUUUUGGACGUCGUUUGAUAGUAGAUCCCAAUACAUUAUAAGUAC----------------- ..............--...((((((...((..(((((.((((.(((...(--(...-.))..))))))).))....)))..)).)))))).............----------------- ( -17.80, z-score = -0.52, R) >droMoj3.scaffold_6496 19515446 99 + 26866924 CUAAAUCAAUAUAU--AAAGUUGGGGUUUUCGCUAACAACGAUCCGCAUA--UGAGUAAUUUUGGACGUCGUUUGAUAGUAGGUCCCAAUACAUUCAAAAUAU----------------- ..............--...((((((..((..((((.((((((((((...(--(.....))..))))..))).))).))))))..)))))).............----------------- ( -18.60, z-score = -0.72, R) >droGri2.scaffold_15245 5259548 101 - 18325388 CUAGAAUAAAAUGU--CAAGUUGGGUUUUUCGCUAACAGCGUACCGCAUAUUUGAAAAAUUUUGGACGUCGUUUGAUAGUAGAUCCCAAUACAUUAUAAUAAU----------------- .........(((((--...((((((...((..(((((.((((.(((.....((....))...))))))).))....)))..)).)))))))))))........----------------- ( -18.90, z-score = -0.91, R) >consensus _CAAAAUUA_AAAU__CCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAA__AUAAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACA____GAAUUU_________________ ...................((((((...((..(((((.((((.(((................))))))).))....)))..)).)))))).............................. (-15.06 = -13.36 + -1.70)

| Location | 6,424,259 – 6,424,359 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.22 |
| Shannon entropy | 0.58109 |
| G+C content | 0.35508 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -14.73 |
| Energy contribution | -13.18 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.45 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
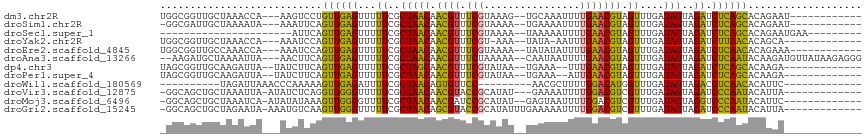
>dm3.chr2R 6424259 100 - 21146708 UGGCGGUUGCUAAACCA---AAGUCCUGUUGAGUUUUUCGCUAACAACGUUUCGUAAAG--UGCAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAGAAU------------ ....((((....)))).---......(((((((...((..(((((.((((((...((((--(....))))))))))).))....)))..)).)))))))......------------ ( -21.20, z-score = -0.17, R) >droSim1.chr2R 5055343 99 - 19596830 -GGCGAUUGCUAAAAUA---AAAUUCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAA--UGAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAGAAU------------ -(((....)))......---.......((((((...((..(((((.((((((...((((--(....))))))))))).))....)))..)).)))))).......------------ ( -19.40, z-score = -0.78, R) >droSec1.super_1 4035797 84 - 14215200 ----------------------AUUCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAA--UAAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAGAAUGAA--------- ----------------------.....((((((...((..(((((.((((((...((((--(....))))))))))).))....)))..)).))))))..........--------- ( -15.80, z-score = -0.91, R) >droYak2.chr2R 18422682 97 + 21139217 UGGCGGUUGCUAAACCA---AAAUCCAGUUGAGUUUUUCGCUAACAACGUUUCGU-AAA--UAUA-AAUUUGAACGUAGUUUGAUAGUAGAUCUUAACACAGCA------------- ..((((((....)))).---.......((((((...((..(((((.((((((...-(((--(...-.)))))))))).))....)))..)).))))))...)).------------- ( -19.30, z-score = -0.82, R) >droEre2.scaffold_4845 3261652 100 + 22589142 UGGCGGUUGCCAAACCA---AAAUCCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAA--UAUAUAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAGAAA------------ ((((....)))).....---.......((((((...((..(((((.((((((...((((--(....))))))))))).))....)))..)).)))))).......------------ ( -20.40, z-score = -1.35, R) >droAna3.scaffold_13266 17899495 110 - 19884421 --AAGAUGCUAAAAUUA---AACUUCAGUUGAGUUUUUCGCUAACAACGUUUCUAAAAA--CAAUAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAAUACAAGAUGUUAUAAGAGGG --.((((.(((..((((---((((...(((.(((.....)))))).((((((..(((..--......))).))))))))))))))..)))))))....................... ( -15.50, z-score = 0.60, R) >dp4.chr3 3599234 98 + 19779522 UAGCGGUUGCAAGAUUA--UAUCUUCAGUUGAGUUUUUCGCUAGCAACGUUUCGUAUAA--UGAAA--UUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAAGA------------- .......((.(((((..--.)))))))((((((...((..(((..(((....(((.(((--.....--.))).)))..)))...)))..)).))))))......------------- ( -17.10, z-score = 0.78, R) >droPer1.super_4 584496 98 - 7162766 UAGCGGUUGCAAGAUUA--UAUCUUCAGUUGAGUUUUUCGCUAACAACGUUUCGUAUAA--UGAAA--AUUGAACGUAGUUUGAUAGUAGAUCUCAGCACAAGA------------- .......((.(((((..--.)))))))((((((...((..(((.((((....(((.(((--(....--)))).)))..).))).)))..)).))))))......------------- ( -19.60, z-score = -0.41, R) >droWil1.scaffold_180569 328365 85 - 1405142 ----------UAGAUUAAACCCAAAAAGUUGAGAUUUUCGCUAACAGUGUUCC---------AACGCUUUUGGACAUGGUUUGAUAGUAGACCUCAACACAUUC------------- ----------..........((((((.((((.((....(((.....))).)))---------)))..))))))....((((((....))))))...........------------- ( -16.90, z-score = -1.16, R) >droVir3.scaffold_12875 16410585 99 - 20611582 -GGCAGCUGCUAAAUUA-AUAUCUCAGGUUGGGUUUUUCGCUAACAACGUACCGCAUAU---GAAAAUUUUGGACGUCGUUUGAUAGUAGAUCCCAAUACAUUA------------- -(((....)))......-.........((((((...((..(((((.((((.(((...((---....))..))))))).))....)))..)).))))))......------------- ( -19.70, z-score = -0.22, R) >droMoj3.scaffold_6496 19515453 100 + 26866924 -GGCAGCUGCUAAAUCA-AUAUAUAAAGUUGGGGUUUUCGCUAACAACGAUCCGCAUAU--GAGUAAUUUUGGACGUCGUUUGAUAGUAGGUCCCAAUACAUUC------------- -((.(.((((((..(((-.........(((((.(....).)))))(((((((((...((--.....))..))))..)))))))))))))).).)).........------------- ( -21.20, z-score = -0.42, R) >droGri2.scaffold_15245 5259555 102 - 18325388 -GGCAGCUGCUAGAAUA-AAAUGUCAAGUUGGGUUUUUCGCUAACAGCGUACCGCAUAUUUGAAAAAUUUUGGACGUCGUUUGAUAGUAGAUCCCAAUACAUUA------------- -((...(((((.(..((-((((.(((((((((((....(((.....))).))).)).))))))...))))))..)(((....))))))))...)).........------------- ( -22.20, z-score = -0.57, R) >consensus _GGCGGUUGCUAAAUUA___AAAUCCAGUUGAGUUUUUCGCUAACAACGUUUCGUAAAA__UAAAAAUUUUGAACGUAGUUUGAUAGUAGAUCUCAACACAGAA_____________ ...........................((((((...((..(((((.((((.(((................))))))).))....)))..)).))))))................... (-14.73 = -13.18 + -1.55)
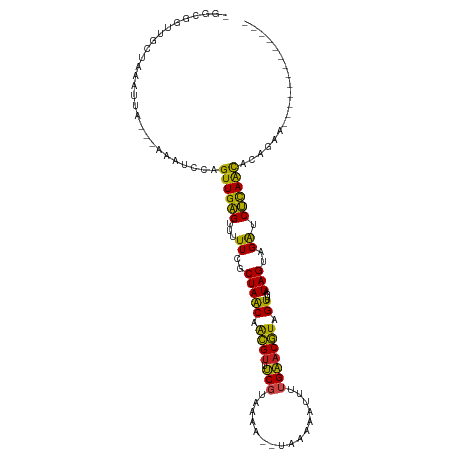
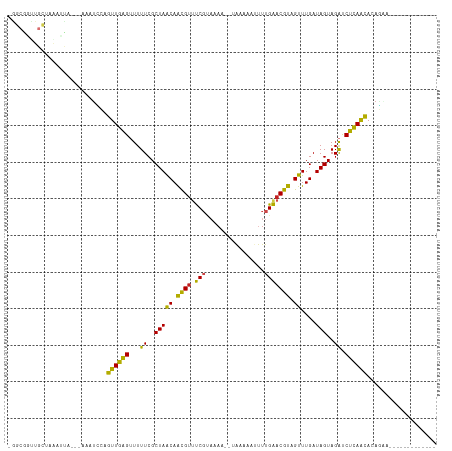
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:33 2011