| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,379,516 – 6,379,607 |
| Length | 91 |
| Max. P | 0.935757 |

| Location | 6,379,516 – 6,379,607 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.79 |
| Shannon entropy | 0.58419 |
| G+C content | 0.56921 |
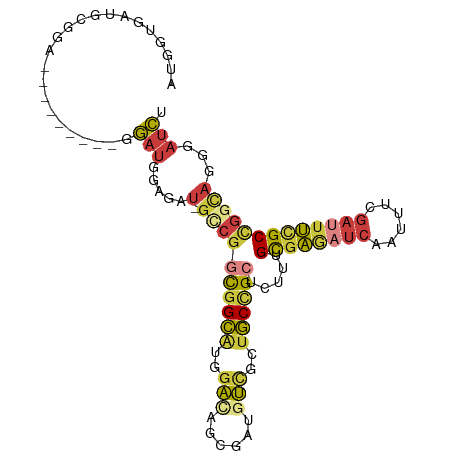
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -19.01 |
| Energy contribution | -19.49 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
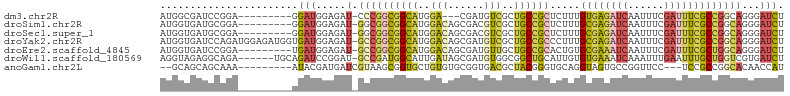
>dm3.chr2R 6379516 91 - 21146708 AUGGCGAUCCGGA---------GGAUGGAGAU-CCCGGCGGCAUGGA---CGAUGUCGCUGCCGCUCUUUGUGAGAUCAAUUUCGAUUUCGCCGGCAGGGAUCU ....(.((((...---------)))).)((((-(((((((((((...---..))))))))((((......((((((((......)))))))))))).))))))) ( -40.10, z-score = -1.91, R) >droSim1.chr2R 5008033 94 - 19596830 AUGGUGAUGCGGA---------GGAUGGAGAU-GGCGGCGGCAUGGACAGCGACGUCGCUGCCGCUCUUUGCGAGAUCAAUUUCGAUUUCGCCGGCAGGGAUCU ..(((..(((.(.---------....((((.(-(((((((((.(.(....).).))))))))))))))..((((((((......))))))))).)))...))). ( -37.80, z-score = -1.29, R) >droSec1.super_1 3991327 94 - 14215200 AUGGUGAUGCGGA---------GGAUGGAGAU-GGCGGCGGCAUGGACAGCGACGUCGCUGCCGCUCUUUGCGAGAUCAAUUUCGAUUUCGCCGGCAGGGAUCU ..(((..(((.(.---------....((((.(-(((((((((.(.(....).).))))))))))))))..((((((((......))))))))).)))...))). ( -37.80, z-score = -1.29, R) >droYak2.chr2R 18378278 103 + 21139217 AUGGUGAUCCAGAUGGAGAUGGUGAUGGAGAU-GCCGGCGGCAUGGACAGCGAUGUCGCUGCCGCCCUUUGCGAGAUCAAUUUCGAUUUCGCCGGCAGGGAUCU .....(((((.....................(-((((((((((..((((....))))..)))))))....((((((((......)))))))).)))).))))). ( -41.35, z-score = -1.36, R) >droEre2.scaffold_4845 3216193 94 + 22589142 AUGGUGAUCCGGA---------UGAUGGAGAU-GCCGGCGGCAUGGACAGCGAUGUUGCUGCCGCACUGUGCGAAAUCAAUUUCGAUUUCGCUGGCAGGGAUCU .....(((((...---------.........(-((.(((((((...(((....)))))))))))))((((((((((((......))))))))..))))))))). ( -38.00, z-score = -2.02, R) >droWil1.scaffold_180569 272454 97 - 1405142 AGGUAGAGGCAGA------UGCAGAUCCGGAU-GCCGAUGGCAUUGAUAGCGAUGUGGCGGCUGCAUUGUGUGAAAUCAAAUUUGAAUUUGCUGGUCGUGAUCU ((((...(((...------.((((((.(((((-((((.(.((((((....)))))).)))))....(((........)))))))).))))))..)))...)))) ( -27.40, z-score = 0.07, R) >anoGam1.chr2L 36429575 90 + 48795086 --GCAGCAGCAAA---------AUACGAUGAUCGUAAGCGGUGCUGUGUGCGGUGACGCUACGGGUGCAGGUAGUGCCGGUUCC---UCCGCCGGCACAACCAU --(((((((((..---------.((((.....)))).....))))))(((((....))).))...))).(((.((((((((...---...)))))))).))).. ( -42.60, z-score = -2.95, R) >consensus AUGGUGAUGCGGA_________GGAUGGAGAU_GCCGGCGGCAUGGACAGCGAUGUCGCUGCCGCUCUUUGCGAGAUCAAUUUCGAUUUCGCCGGCAGGGAUCU .................................((((((((((..(((......)))..)))))).....((((((((......))))))))))))........ (-19.01 = -19.49 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:28 2011