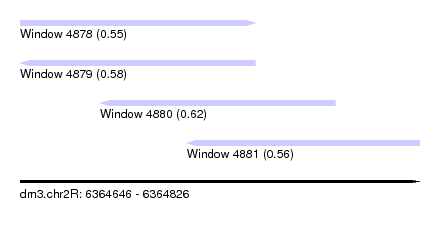
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,364,646 – 6,364,826 |
| Length | 180 |
| Max. P | 0.621474 |

| Location | 6,364,646 – 6,364,752 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.09 |
| Shannon entropy | 0.59380 |
| G+C content | 0.39757 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -10.73 |
| Energy contribution | -12.72 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6364646 106 + 21146708 ----GCGCAACGAAAAUUGAUAAAGCGAGCGGGGUAUCUUUGAGAUACUCGACUACAAGAUAUCCACUAUUACAUGGAU-UCACAUGAGUUAUUGAACUAAAAGUGCUGUG ----((((..((....(((......))).))(((((((((((((...))))).....)))))))).......((((...-...))))((((....))))....)))).... ( -22.00, z-score = -0.81, R) >droSim1.chr2R 4993245 106 + 19596830 ----GCGCAACUAAAAUUGAUAAAGCGAGCAGGGUAUCUUUGAGAUACUCGACUACAAGAUACCCGCUAUUACAUGGAU-UCACGUGGGUUAUUGAACUAAAAGUGCUUUG ----((((........(..((((....(((.(((((((((((((...))))).....)))))))))))....((((...-...))))..))))..).......)))).... ( -27.06, z-score = -2.27, R) >droSec1.super_1 3976282 107 + 14215200 ----GCGCAACGAAAAUUGAUAAAGCGAGCGGGGUAUCUUUGAGAUACUCGACUACAAGAUACCCGCUAUUACAUGGAUAUCACGUGGGUUAUUGAACUAAAAGUGCUUUG ----((((........(..((((....((((((.((((((((((...))))).....)))))))))))....((((.......))))..))))..).......)))).... ( -26.56, z-score = -1.78, R) >droAna3.scaffold_13266 17836919 87 + 19884421 AAUAAAAAAAAGCAAAUAAACUGAAGAGGAGAAGGAGGUGUGAGCAGCUCGACUUUGAGAUACCUUUGAUAAGCUAAGCUCUAUUCA------------------------ ......................(((..((((..((((((((......((((....))))))))))))...........)))).))).------------------------ ( -12.32, z-score = 1.25, R) >consensus ____GCGCAACGAAAAUUGAUAAAGCGAGCGGGGUAUCUUUGAGAUACUCGACUACAAGAUACCCGCUAUUACAUGGAU_UCACGUGGGUUAUUGAACUAAAAGUGCUUUG ....((((...................(((.(((((((((((((...))))).....)))))))))))....((((.......))))................)))).... (-10.73 = -12.72 + 2.00)


| Location | 6,364,646 – 6,364,752 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 63.09 |
| Shannon entropy | 0.59380 |
| G+C content | 0.39757 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -9.75 |
| Energy contribution | -11.75 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6364646 106 - 21146708 CACAGCACUUUUAGUUCAAUAACUCAUGUGA-AUCCAUGUAAUAGUGGAUAUCUUGUAGUCGAGUAUCUCAAAGAUACCCCGCUCGCUUUAUCAAUUUUCGUUGCGC---- ..((((......((((....))))...((((-((((((......))))))........((.(.(((((.....))))).).)))))).............))))...---- ( -18.00, z-score = -0.14, R) >droSim1.chr2R 4993245 106 - 19596830 CAAAGCACUUUUAGUUCAAUAACCCACGUGA-AUCCAUGUAAUAGCGGGUAUCUUGUAGUCGAGUAUCUCAAAGAUACCCUGCUCGCUUUAUCAAUUUUAGUUGCGC---- .(((((...................(((((.-...)))))...((((((((((((..((........))..))))))))).))).))))).................---- ( -24.80, z-score = -2.32, R) >droSec1.super_1 3976282 107 - 14215200 CAAAGCACUUUUAGUUCAAUAACCCACGUGAUAUCCAUGUAAUAGCGGGUAUCUUGUAGUCGAGUAUCUCAAAGAUACCCCGCUCGCUUUAUCAAUUUUCGUUGCGC---- .(((((...................(((((.....)))))...((((((((((((..((........))..)))))).)))))).))))).................---- ( -24.80, z-score = -2.45, R) >droAna3.scaffold_13266 17836919 87 - 19884421 ------------------------UGAAUAGAGCUUAGCUUAUCAAAGGUAUCUCAAAGUCGAGCUGCUCACACCUCCUUCUCCUCUUCAGUUUAUUUGCUUUUUUUUAUU ------------------------......((((..(((((..(..............)..)))))))))......................................... ( -8.94, z-score = 1.57, R) >consensus CAAAGCACUUUUAGUUCAAUAACCCACGUGA_AUCCAUGUAAUAGCGGGUAUCUUGUAGUCGAGUAUCUCAAAGAUACCCCGCUCGCUUUAUCAAUUUUCGUUGCGC____ .(((((...................(((((.....)))))...((((((((((((..((........))..))))))))).))).)))))..................... ( -9.75 = -11.75 + 2.00)

| Location | 6,364,682 – 6,364,788 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.87 |
| Shannon entropy | 0.62958 |
| G+C content | 0.36956 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -10.25 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6364682 106 - 21146708 UUACGUUUUUAAGUGUCGCAGUACUUCAGAAAUGUU----------CACAGCACUUUUAGUUCAAUAACUCAUGUGA-AUCCAUGUAAUAGUGGAUAUCUUGUAGUCGAGUAUCUCA ..(((((((((((((......))))).))))))))(----------((((......(((......)))....)))))-((((((......))))))..((((....))))....... ( -18.20, z-score = 0.11, R) >droSim1.chr2R 4993281 106 - 19596830 UUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU----------CAAAGCACUUUUAGUUCAAUAACCCACGUGA-AUCCAUGUAAUAGCGGGUAUCUUGUAGUCGAGUAUCUCA ..((((((((((((((....)))))).)))))))).----------....((.......(((....)))..(((((.-...)))))....))(((...((((....))))...))). ( -21.60, z-score = -0.84, R) >droSec1.super_1 3976318 107 - 14215200 UUGCGUUUUUAAGUGCGGCCGUACUUAAGAAAUGUU----------CAAAGCACUUUUAGUUCAAUAACCCACGUGAUAUCCAUGUAAUAGCGGGUAUCUUGUAGUCGAGUAUCUCA ..((.(((((((((((....))))))))))).....----------.........................(((((.....)))))....))(((...((((....))))...))). ( -22.20, z-score = -0.56, R) >droYak2.chr2R 18363771 116 + 21139217 UUACGUUUUUGAGUGCGAUAGUACUUCAGAAAUGUUUUCAUUCCUACAAAUUACUUUCAGUGCAGAAAUCUACUUGC-AUCAAUGUAAAAGCGGGUAUCUUGUAGUCGAGUAUCUCA ..((((((((((((((....)))).))))))))))....((((((((((..(((((...((((((........))))-))....((....)))))))..))))))..))))...... ( -27.70, z-score = -2.10, R) >droEre2.scaffold_4845 3201529 102 + 22589142 UUACGUUUUUAAGUGCAACAGCACUUCAGAAAUGUUUUCAUUGCUUUACGGUGCUUUCAGUGCAGAAACCUACU---------------AGCGGCUAUCUUGUAGUCGAGCAUCUCG ..((((((((((((((....)))))).))))))))..............((((((...((((........))))---------------..(((((((...)))))))))))))... ( -26.70, z-score = -1.59, R) >droAna3.scaffold_13266 17836959 86 - 19884421 UUUUGGCUUUAGGUAUAAAGAUAU-----AAGU-------------UAUAAUGUAUUU-GAAUAGAGCUUAGCUU------------AUCAAAGGUAUCUCAAAGUCGAGCUGCUCA .(((((((((((((((......((-----((((-------------((...(.(((..-..))).)...))))))------------)).....)))))).)))))))))....... ( -18.90, z-score = -1.06, R) >consensus UUACGUUUUUAAGUGCGACAGUACUUCAGAAAUGUU__________CAAAGCACUUUUAGUUCAAAAACCCACGUGA_AUCCAUGUAAUAGCGGGUAUCUUGUAGUCGAGUAUCUCA ..((((((((((((((....)))))).)))))))).........................................................(((...((((....))))...))). (-10.25 = -11.12 + 0.87)

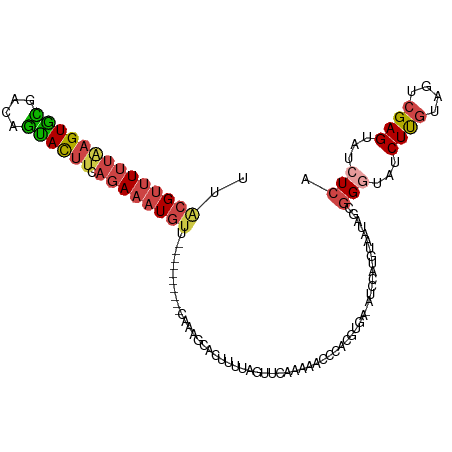
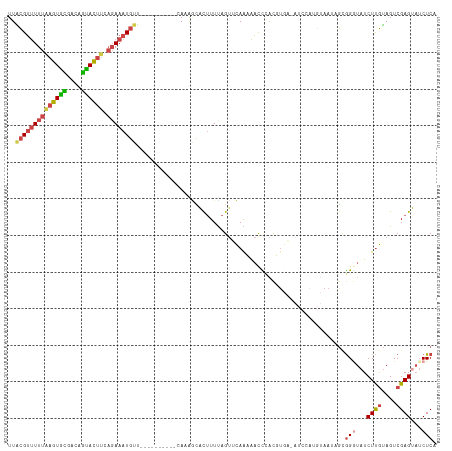
| Location | 6,364,721 – 6,364,826 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Shannon entropy | 0.35682 |
| G+C content | 0.39357 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -16.76 |
| Energy contribution | -15.40 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6364721 105 - 21146708 AUGACCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGUCGCAGUACUUCAGAAAUGUU----------CACAGCACUUUUAGUUCAAUAACUCAUGUGA ..(((.((((..((((..((((((........))))))...))))...)))))))...((....((....))..----------.))..(((....((((....))))...))). ( -18.90, z-score = -0.10, R) >droSim1.chr2R 4993320 105 - 19596830 AUGACCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU----------CAAAGCACUUUUAGUUCAAUAACCCACGUGA ......(((((.(((...((((((........))))))....((((((((((((....)))))).)))))))))----------.)))))......................... ( -22.20, z-score = -1.06, R) >droSec1.super_1 3976358 105 - 14215200 AUGCCCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUGCGUUUUUAAGUGCGGCCGUACUUAAGAAAUGUU----------CAAAGCACUUUUAGUUCAAUAACCCACGUGA ......(((((.((((..((((((........)))))).....(((((((((((....))))))))))).))))----------.)))))......................... ( -24.70, z-score = -1.63, R) >droYak2.chr2R 18363810 112 + 21139217 ---ACCGUUUCUAACGACCAUACUCAACCAAAAGUGUGUUACGUUUUUGAGUGCGAUAGUACUUCAGAAAUGUUUUCAUUCCUACAAAUUACUUUCAGUGCAGAAAUCUACUUGC ---...((((((.((...((((((........))))))..((((((((((((((....)))).))))))))))........................))..))))))........ ( -22.90, z-score = -1.72, R) >droEre2.scaffold_4845 3201557 112 + 22589142 AUGACCGUUUCUAACGACCAUGCUCAACCAAAAGUGUGUUACGUUUUUAAGUGCAACAGCACUUCAGAAAUGUUUUCAUUGCUUUACGGUGCUUUCAGUGCAGAAACCUACU--- ......((((((.((......((...(((.((((((((..((((((((((((((....)))))).))))))))...)).))))))..))))).....))..)))))).....--- ( -27.60, z-score = -1.63, R) >consensus AUGACCGCUUUUAACGACCACACUCAACCAAUAGUGUGUUACGUUUUUAAGUGCGGCAGUACUUCAGAAAUGUU__________CAAAGCACUUUUAGUUCAAUAACCCACGUGA ......(((.........((((((........))))))..((((((((((((((....)))))).))))))))..............)))......................... (-16.76 = -15.40 + -1.36)

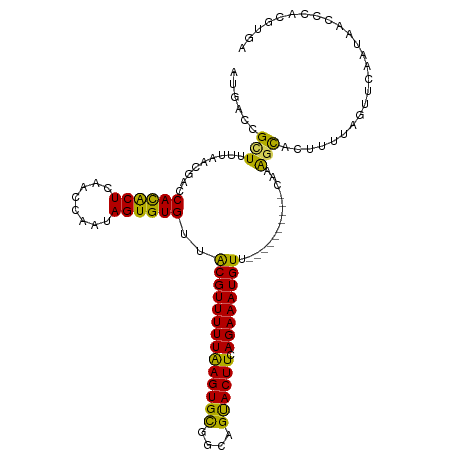
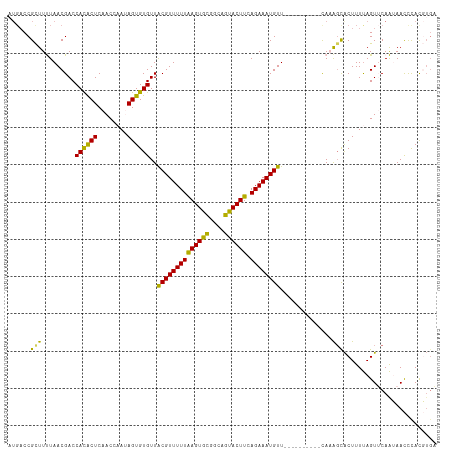
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:26 2011