
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,339,652 – 6,339,747 |
| Length | 95 |
| Max. P | 0.900992 |

| Location | 6,339,652 – 6,339,747 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 65.52 |
| Shannon entropy | 0.73276 |
| G+C content | 0.55148 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.66 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
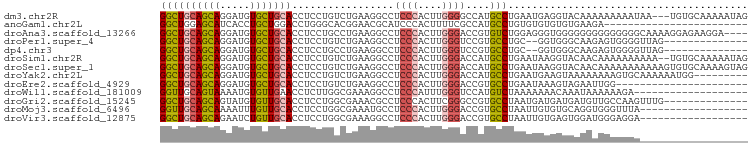
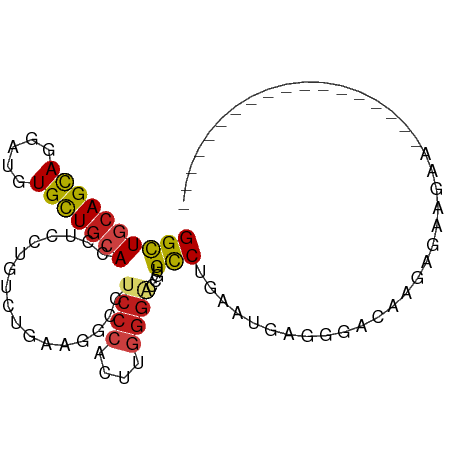
>dm3.chr2R 6339652 95 + 21146708 GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGGCCAUGCCUGAAUGAGGUACAAAAAAAAAUAA---UGUGCAAAAAUAG (((..((((((((.(((...))).))))).)))..((((((......))))))..)))........(((((...........---)))))........ ( -31.10, z-score = -1.83, R) >anoGam1.chr2L 28902585 74 - 48795086 GGCUGGAGCAUCACCUGCUGGACCUGGGCACGGAACGCAUCCCACUUUUCGCCAUGCCUGUGUGUGUGUGAAGA------------------------ ....(((((....((((((.......)))).))...)).)))...((((((((((((....))))).)))))))------------------------ ( -18.20, z-score = 2.04, R) >droAna3.scaffold_13266 17809903 94 + 19884421 GGCUGCAGCAGGAUGUGCUGCACCUCCUGCCUGAAGGCCUCCCACUUGGGACCGUGUCUGGAGGGUGGGGGGGGGGGGGGCAAAAGGAGAAGGA---- .(((((((((.....)))))).(((((((((....)))(((((((((.((((...))))...))))))))))))))).))).............---- ( -43.00, z-score = -0.77, R) >droPer1.super_4 500767 82 + 7162766 GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGUCCGUGCCUGC--GGUGGGCAAGAGUGGGGUUAG-------------- ((((.((((((((.(((...))).))))).)))..))))((((((((..((((..(((...--)))))))..))))))))....-------------- ( -38.80, z-score = -1.42, R) >dp4.chr3 3515343 82 - 19779522 GGCUGCAGCAGGAUGUGCUGCACCUCCUGCCUGAAGGCCUCCCACUUGGGUCCGUGCCUGC--GGUGGGCAAGAGUGGGGUUAG-------------- ((((.((((((((.(((...))).))))).)))..))))((((((((..((((..(((...--)))))))..))))))))....-------------- ( -38.80, z-score = -1.01, R) >droSim1.chr2R 4968010 96 + 19596830 GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCAUGCCUGAAUAAGGUACAACAAAAAAAAAA--UGUGCAAAAAUAG ((((.((((((((.(((...))).))))).)))..))))((((....))))...(((((.....)))))..............--............. ( -29.00, z-score = -2.01, R) >droSec1.super_1 3951387 98 + 14215200 GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCAUGCCUGAAUAAGGUACAACAAAAAAAAAAAGUGUGCAAAAGUAG ((((.((((((((.(((...))).))))).)))..))))((((....))))...(((((.....)))))............................. ( -29.00, z-score = -1.25, R) >droYak2.chr2L 18976967 89 + 22324452 GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCAUGCCUGAAUGAAGUAAAAAAAAGUGCAAAAAAUGG--------- ((((.((((((((.(((...))).))))).)))..))))((((....))))((((((((................)).))).....)))--------- ( -26.39, z-score = -1.22, R) >droEre2.scaffold_4929 18599782 76 - 26641161 GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCGUGCCUGAAUAAAGUAGAAUUGG---------------------- ((.(((((((.....))))))))).((..((((.(((((((((....))))..).))))........))))...))---------------------- ( -26.00, z-score = -1.17, R) >droWil1.scaffold_181009 1128883 79 - 3585778 GGUUGCAGUAAAAUGUGUUGAACCUCUUGGCGAAAGGCCUCCCAUUUGGGUCCAUGUCUAAAAAAACAAAUAAAAAAGA------------------- (((((((......)))....))))(((((((.....))).(((....))).........................))))------------------- ( -11.60, z-score = 0.93, R) >droGri2.scaffold_15245 5158785 84 + 18325388 GGCUGCAGCAGUAUGUGUUGCACCUCCUGGCGAAACGCCUCCCACUUCGGGCCGUGCCUAAUGAUGAUGAUGUUGCCAAGUUUG-------------- ((((((((((.....)))))))......((((....((((........))))..))))................))).......-------------- ( -22.50, z-score = 0.39, R) >droMoj3.scaffold_6496 19405426 80 - 26866924 GGUUGCAGCAAAAUUUGUUGCACCUCCUGGCGAAAUGCCUCCCACUUGGGACCGUGCCUAAUUGUGUGCAGGUGGGUUUA------------------ (((.((((((.....)))))))))....(((.....))).((((((((.(..((........))..).))))))))....------------------ ( -27.40, z-score = -0.92, R) >droVir3.scaffold_12875 16292293 80 + 20611582 GGCUGCAGCAGAAUCUGUUGCACCUCCUGGCGAAAGGCCUCCCACUUGGGACCGUGCCUAAUUGUGAGUGGAUGGGAGGA------------------ ...((((((((...))))))))(((((((((.....)))..(((((((((......)))......))))))..)))))).------------------ ( -32.10, z-score = -1.51, R) >consensus GGCUGCAGCAGGAUGUGCUGCACCUCCUGUCUGAAGGCCUCCCACUUGGGACCGUGCCUGAAUGAGGGACAAGAGAAGAA__________________ ((((((((((.....))))))).................((((....))))....)))........................................ (-14.56 = -14.66 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:21 2011