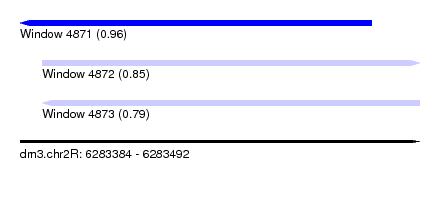
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,283,384 – 6,283,492 |
| Length | 108 |
| Max. P | 0.959127 |

| Location | 6,283,384 – 6,283,479 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.22 |
| Shannon entropy | 0.62943 |
| G+C content | 0.43264 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -11.03 |
| Energy contribution | -11.00 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
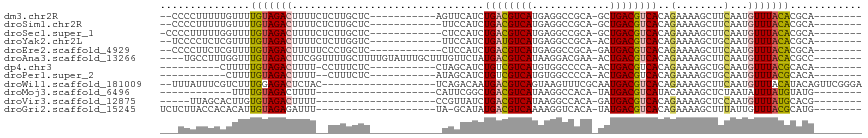
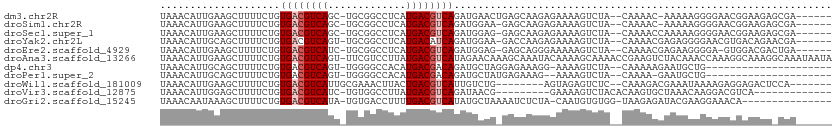
>dm3.chr2R 6283384 95 - 21146708 --CCCCUUUUUGUUUUGUAGACUUUUCUCUUGCUC-----------AGUUCAUCUGACGUCAUGAGGCCGCA-GCUGACGUCACAGAAAAGCUUCAAUGUUUACACGCA-------- --........(((..(((((((.........(((.-----------..(((...((((((((....((....-))))))))))..))).)))......))))))).)))-------- ( -20.56, z-score = -0.41, R) >droSim1.chr2R 4906873 94 - 19596830 --CCCCUUUUUGUUUUGUAGACUUUUCUCUUGCUC------------UUCCAUCUGACGUCAUGAGGCCGCA-GCUGACGUCACAGAAAAGCUUCAAUGUUUACACGCA-------- --........(((..(((((((.........(((.------------(((....((((((((....((....-))))))))))..))).)))......))))))).)))-------- ( -21.16, z-score = -1.17, R) >droSec1.super_1 3896077 95 - 14215200 -CCCCUUUUUGGUUUUGUAGACUUUUCUCUUGCUC------------CUCCAUCUGACGUCAUGAGGCCGCA-GCUGACGUCACAGAAAAGCUUCAAUGUUUACACGCA-------- -..((.....))...(((((((.........(((.------------.((....((((((((....((....-))))))))))..))..)))......)))))))....-------- ( -19.96, z-score = -0.35, R) >droYak2.chr2L 18909566 94 - 22324452 --UCCCCUCUCGUUUUGUAGACUUUUCUCUUGGUC------------UUCCAUCUGAUGUCAUGAGGCCGCA-ACUGACGUCACAGAAAAGCUGCAAUGUUUACACGCA-------- --............((((((.(((((((..(((..------------..)))..((((((((((......))-..)))))))).)))))))))))))............-------- ( -23.00, z-score = -1.30, R) >droEre2.scaffold_4929 18550586 94 + 26641161 --CCCCUUCUCGUUUUGUAGACUUUUUCCCUGCUC------------CUCCAUCUGACGUCAUGAGGCCGCA-GAUGACGUCACAGAAAAGCUUCAAUGUUUACACGCA-------- --.........((..(((((((.........(((.------------.((....(((((((((..(....).-.)))))))))..))..)))......))))))).)).-------- ( -20.76, z-score = -1.30, R) >droAna3.scaffold_13266 4746266 104 - 19884421 ----UGCCUUUGGUUUGUAGACUUCGGUUUUGCUUUUGUAUUUGCUUUGUUCUAUGACGUCAUAAGGACGAA-ACUGACGUCACAGAAAAGCUUCAAUGUUUACACGCC-------- ----.......(((.(((((((........(((....)))...(((((..(((.((((((((..........-..)))))))).))))))))......))))))).)))-------- ( -27.50, z-score = -1.85, R) >dp4.chr3 12616189 86 + 19779522 ----------CUUUUUGUAGACUUUU-CCUUUCUC-----------CUAGCAUCUGUCGUCAUGUGGCCCCA-ACUGACGUCACAGAAAAGCUGCAAUGUUUACGCACA-------- ----------....((((((.(((((-(((.....-----------..))....((.(((((..((....))-..))))).))..))))))))))))............-------- ( -19.00, z-score = -1.42, R) >droPer1.super_2 1673501 84 + 9036312 -----------CUUUUGUAGACUUUU--CUUUCUC-----------AUAGCAUCUGUCGUCAUGUGGCCCCA-ACUGACGUCACAGAAAAGCUGCAAUGUUUACGCACA-------- -----------...((((((.(((((--((.....-----------........((.(((((..((....))-..))))).)).)))))))))))))............-------- ( -19.64, z-score = -1.44, R) >droWil1.scaffold_181009 1076823 96 + 3585778 --UUUAUUUCGUCUUUGGAGACUCUAC-------------------UCAGACAAUGACGUCAGUAAGUUUCGCAAUGACGUCACAGAAAAGCUUCAAUGUUUACAUACAGUUCGGGA --........((((....))))....(-------------------((..((..((((((((.............)))))))).....((((......)))).......))..))). ( -19.72, z-score = -0.27, R) >droMoj3.scaffold_6496 19335024 76 + 26866924 ------------UUUUGUAGACUUUU--------------------CAUUCGGCUGACGUCAUAAGGCCACA-UAUGACGUCAUACAAAAGCUCUAAUAUUUAUGUAUG-------- ------------.....(((((((((--------------------.....(..((((((((((.(....).-))))))))))..)))))).)))).............-------- ( -18.10, z-score = -2.88, R) >droVir3.scaffold_12875 16225235 83 - 20611582 -----UUAGCACUUGUGUAGACUUUU--------------------CCGUUAUCUGACGUCAUAAGGCCACA-GAUGACGUCACAGAAAAGCUCCAAUGUUUAUGCACG-------- -----...(((....((.((.(((((--------------------((((((((((..(((....)))..))-))))))).....)))))))).)).......)))...-------- ( -21.20, z-score = -1.98, R) >droGri2.scaffold_15245 13058625 87 - 18325388 UCUCUUACCACACAUUGUAGAGAUUU--------------------UA-GCAUAUGACGUCAAAAGGUCACA-UAUGACGUCACAGAAAAGCUUUAUUGUUUACGCAUG-------- (((((.((........)))))))...--------------------..-((...((((((((...(....).-..)))))))).....((((......))))..))...-------- ( -18.00, z-score = -1.72, R) >consensus ___CCCUUUUCGUUUUGUAGACUUUU__CUUGCUC____________UUCCAUCUGACGUCAUGAGGCCGCA_ACUGACGUCACAGAAAAGCUUCAAUGUUUACACACA________ ...............(((((((................................((((((((.............))))))))..(........)...)))))))............ (-11.03 = -11.00 + -0.03)
| Location | 6,283,390 – 6,283,492 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 65.31 |
| Shannon entropy | 0.73919 |
| G+C content | 0.42924 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.09 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.846776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
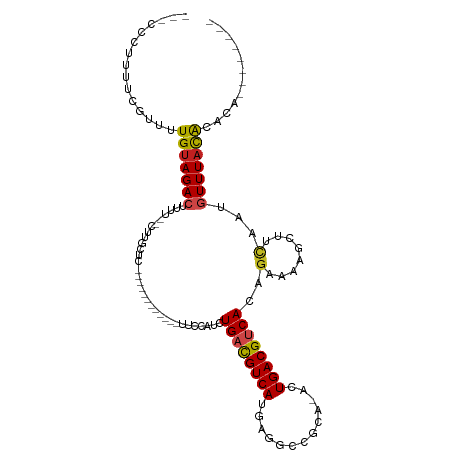
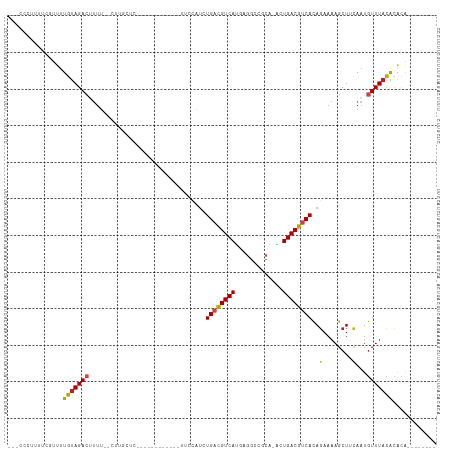
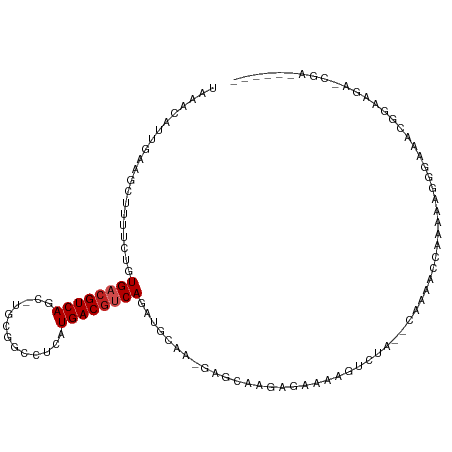
>dm3.chr2R 6283390 102 + 21146708 UAAACAUUGAAGCUUUUCUGUGACGUCAGC-UGCGGCCUCAUGACGUCAGAUGAACUGAGCAAGAGAAAAGUCUA--CAAAAC-AAAAAGGGGAACGGAAGAGCGA------ ...........(((((((((((((((((((-....))....)))))))((((...((.......))....)))).--......-..........))))))))))..------ ( -26.10, z-score = -1.13, R) >droSim1.chr2R 4906879 101 + 19596830 UAAACAUUGAAGCUUUUCUGUGACGUCAGC-UGCGGCCUCAUGACGUCAGAUGGAA-GAGCAAGAGAAAAGUCUA--CAAAAC-AAAAAGGGGAACGGAAGAGCGA------ ...........(((((((((((((((((((-....))....)))))))...((((.-...(....).....))))--......-..........))))))))))..------ ( -25.40, z-score = -1.08, R) >droSec1.super_1 3896083 102 + 14215200 UAAACAUUGAAGCUUUUCUGUGACGUCAGC-UGCGGCCUCAUGACGUCAGAUGGAG-GAGCAAGAGAAAAGUCUA--CAAAACCAAAAAGGGGAACGGAAGAGCGA------ ...........(((((((((((((.((...-(((..((((.((....))....)))-).)))...))...)))..--.....((......))..))))))))))..------ ( -29.30, z-score = -1.86, R) >droYak2.chr2L 18909572 102 + 22324452 UAAACAUUGCAGCUUUUCUGUGACGUCAGU-UGCGGCCUCAUGACAUCAGAUGGAA-GACCAAGAGAAAAGUCUA--CAAAACGAGAGGGGAACGUGACAGAACGA------ ......(..(((.....)))..)((((.((-..((..(((..(((.((...(((..-..)))...))...)))..--.....(....))))..))..)).).))).------ ( -20.40, z-score = 0.80, R) >droEre2.scaffold_4929 18550592 101 - 26641161 UAAACAUUGAAGCUUUUCUGUGACGUCAUC-UGCGGCCUCAUGACGUCAGAUGGAG-GAGCAGGGAAAAAGUCUA--CAAAACGAGAAGGGGA-GUGGACGACUGA------ .............((..((((..(..((((-((((.........)).))))))..)-..))))..))...(((((--(....(.....)....-))))))......------ ( -28.70, z-score = -1.20, R) >droAna3.scaffold_13266 4746272 111 + 19884421 UAAACAUUGAAGCUUUUCUGUGACGUCAGU-UUCGUCCUUAUGACGUCAUAGAACAAAGCAAAUACAAAAGCAAAACCGAAGUCUACAAACCAAAGGCAAAGGCAAAUAAUA ......(((..(((((((((((((((((..-..........))))))))))))..)))))................((...((((.........))))...)))))...... ( -26.30, z-score = -3.79, R) >dp4.chr3 12616195 87 - 19779522 UAAACAUUGCAGCUUUUCUGUGACGUCAGU-UGGGGCCACAUGACGACAGAUGCUAGGAGAAAGG-AAAAGUCUA--CAAAAAGAAUGCUG--------------------- .........((((..((((.((.(((((..-((....))..)))))..((((.((........))-....)))).--))...)))).))))--------------------- ( -17.90, z-score = 0.03, R) >droPer1.super_2 1673507 85 - 9036312 UAAACAUUGCAGCUUUUCUGUGACGUCAGU-UGGGGCCACAUGACGACAGAUGCUAUGAGAAAG--AAAAGUCUA--CAAAA-GAAUGCUG--------------------- ........(((((...(((((..(((((..-((....))..)))))))))).))).(((((...--.....))).--))...-....))..--------------------- ( -18.00, z-score = -0.26, R) >droWil1.scaffold_181009 1076837 95 - 3585778 UAAACAUUGAAGCUUUUCUGUGACGUCAUUGCGAAACUUACUGACGUCAUUGUCUG--------AGUAGAGUCUC--CAAAGACGAAAUAAAAGAGGAGACUCCA------- ...........((((..(.(((((((((.((.......)).))))))))).)...)--------))).(((((((--(.................))))))))..------- ( -26.83, z-score = -2.41, R) >droVir3.scaffold_12875 16225241 89 + 20611582 UAAACAUUGGAGCUUUUCUGUGACGUCAUC-UGUGGCCUUAUGACGUCAGAUAACG---------GAAAAGUCUACACAAGUGCUAAACAAGGACGUCA------------- .......((((.(((((((((((((((((.-.(....)..)))))))).....)))---------)))))))))).....((.((.....)).))....------------- ( -23.90, z-score = -1.59, R) >droGri2.scaffold_15245 13058631 94 + 18325388 UAAACAAUAAAGCUUUUCUGUGACGUCAUA-UGUGACCUUUUGACGUCAUAUGCUAAAAUCUCUA-CAAUGUGUGG-UAAGAGAUACGAAGGAAACA--------------- ..........(((.....((((((((((..-..........)))))))))).)))...(((((((-(........)-).)))))).....(....).--------------- ( -22.40, z-score = -2.17, R) >consensus UAAACAUUGAAGCUUUUCUGUGACGUCAGC_UGCGGCCUCAUGACGUCAGAUGCAA_GAGCAAGAGAAAAGUCUA__CAAAACCAAAAAGGGAAACGGAAGA_CGA______ ....................((((((((.............))))))))............................................................... ( -8.82 = -9.09 + 0.27)
| Location | 6,283,390 – 6,283,492 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.31 |
| Shannon entropy | 0.73919 |
| G+C content | 0.42924 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -9.55 |
| Energy contribution | -9.65 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.789520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
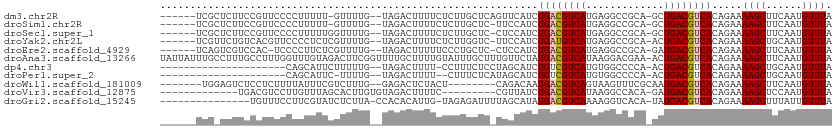
>dm3.chr2R 6283390 102 - 21146708 ------UCGCUCUUCCGUUCCCCUUUUU-GUUUUG--UAGACUUUUCUCUUGCUCAGUUCAUCUGACGUCAUGAGGCCGCA-GCUGACGUCACAGAAAAGCUUCAAUGUUUA ------..(((.(((.((.........(-(..(((--.(((......)))....)))..))...(((((((....((....-))))))))))).))).)))........... ( -17.60, z-score = 0.72, R) >droSim1.chr2R 4906879 101 - 19596830 ------UCGCUCUUCCGUUCCCCUUUUU-GUUUUG--UAGACUUUUCUCUUGCUC-UUCCAUCUGACGUCAUGAGGCCGCA-GCUGACGUCACAGAAAAGCUUCAAUGUUUA ------......................-...(((--.((.(((((((..((...-...))..((((((((....((....-)))))))))).))))))))).)))...... ( -16.80, z-score = 0.50, R) >droSec1.super_1 3896083 102 - 14215200 ------UCGCUCUUCCGUUCCCCUUUUUGGUUUUG--UAGACUUUUCUCUUGCUC-CUCCAUCUGACGUCAUGAGGCCGCA-GCUGACGUCACAGAAAAGCUUCAAUGUUUA ------..(((.(((.((.........(((....(--((((......)).)))..-..)))...(((((((....((....-))))))))))).))).)))........... ( -19.60, z-score = 0.04, R) >droYak2.chr2L 18909572 102 - 22324452 ------UCGUUCUGUCACGUUCCCCUCUCGUUUUG--UAGACUUUUCUCUUGGUC-UUCCAUCUGAUGUCAUGAGGCCGCA-ACUGACGUCACAGAAAAGCUGCAAUGUUUA ------..........................(((--(((.(((((((..(((..-..)))..((((((((((......))-..)))))))).)))))))))))))...... ( -23.00, z-score = -0.82, R) >droEre2.scaffold_4929 18550592 101 + 26641161 ------UCAGUCGUCCAC-UCCCCUUCUCGUUUUG--UAGACUUUUUCCCUGCUC-CUCCAUCUGACGUCAUGAGGCCGCA-GAUGACGUCACAGAAAAGCUUCAAUGUUUA ------......(((.((-...............)--).))).........(((.-.((....(((((((((..(....).-.)))))))))..))..)))........... ( -16.76, z-score = 0.52, R) >droAna3.scaffold_13266 4746272 111 - 19884421 UAUUAUUUGCCUUUGCCUUUGGUUUGUAGACUUCGGUUUUGCUUUUGUAUUUGCUUUGUUCUAUGACGUCAUAAGGACGAA-ACUGACGUCACAGAAAAGCUUCAAUGUUUA ...(((..(((.........)))..)))(((....))).(((....)))...(((((..(((.((((((((..........-..)))))))).))))))))........... ( -23.20, z-score = -0.80, R) >dp4.chr3 12616195 87 + 19779522 ---------------------CAGCAUUCUUUUUG--UAGACUUUU-CCUUUCUCCUAGCAUCUGUCGUCAUGUGGCCCCA-ACUGACGUCACAGAAAAGCUGCAAUGUUUA ---------------------...........(((--(((.(((((-(((.......))....((.(((((..((....))-..))))).))..))))))))))))...... ( -19.00, z-score = -1.49, R) >droPer1.super_2 1673507 85 + 9036312 ---------------------CAGCAUUC-UUUUG--UAGACUUUU--CUUUCUCAUAGCAUCUGUCGUCAUGUGGCCCCA-ACUGACGUCACAGAAAAGCUGCAAUGUUUA ---------------------........-..(((--(((.(((((--((.............((.(((((..((....))-..))))).)).)))))))))))))...... ( -19.64, z-score = -1.52, R) >droWil1.scaffold_181009 1076837 95 + 3585778 -------UGGAGUCUCCUCUUUUAUUUCGUCUUUG--GAGACUCUACU--------CAGACAAUGACGUCAGUAAGUUUCGCAAUGACGUCACAGAAAAGCUUCAAUGUUUA -------((((((((((.................)--)))))))))..--------.(((((.((((((((.............))))))))..(((....)))..))))). ( -28.15, z-score = -2.82, R) >droVir3.scaffold_12875 16225241 89 - 20611582 -------------UGACGUCCUUGUUUAGCACUUGUGUAGACUUUUC---------CGUUAUCUGACGUCAUAAGGCCACA-GAUGACGUCACAGAAAAGCUCCAAUGUUUA -------------(((((.....(((((.((....))))))).....---------)))))..(((((((((..(....).-.))))))))).....((((......)))). ( -20.90, z-score = -1.07, R) >droGri2.scaffold_15245 13058631 94 - 18325388 ---------------UGUUUCCUUCGUAUCUCUUA-CCACACAUUG-UAGAGAUUUUAGCAUAUGACGUCAAAAGGUCACA-UAUGACGUCACAGAAAAGCUUUAUUGUUUA ---------------.......(((((((((((.(-(........)-)))))))....))...((((((((...(....).-..))))))))..)))((((......)))). ( -17.20, z-score = -0.88, R) >consensus ______UCG_UCUUCCGUUUCCCUUUUUCGUUUUG__UAGACUUUUCUCUUGCUC_UUCCAUCUGACGUCAUGAGGCCGCA_ACUGACGUCACAGAAAAGCUUCAAUGUUUA ...............................................................((((((((.............)))))))).....((((......)))). ( -9.55 = -9.65 + 0.10)
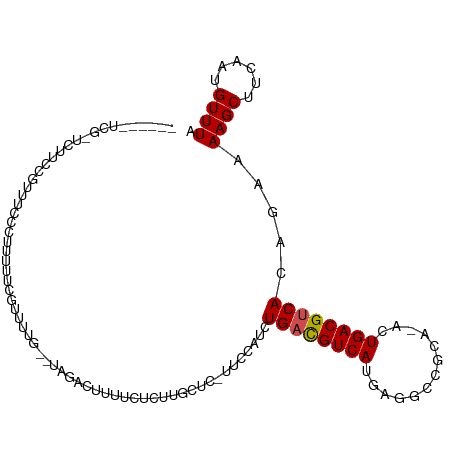
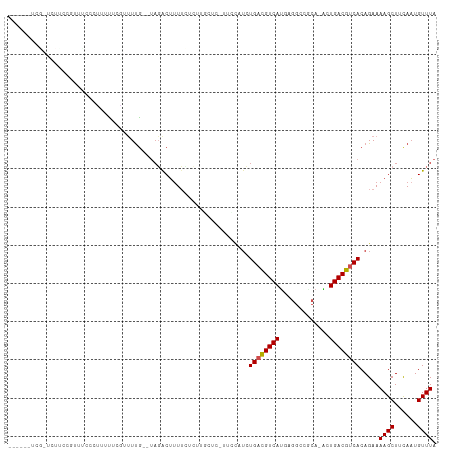
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:19 2011