| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,272,799 – 6,272,907 |
| Length | 108 |
| Max. P | 0.528036 |

| Location | 6,272,799 – 6,272,907 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.56341 |
| G+C content | 0.47497 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -9.95 |
| Energy contribution | -10.36 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
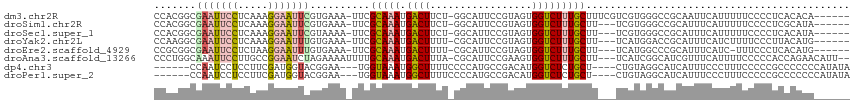
>dm3.chr2R 6272799 108 - 21146708 CCACGGCGAAUUCCUCAAAGGAAUUCGUGAAA-UUCGCAAAUGACUUCU-GGCAUUCCGUAGUGGUCUUUGCUUUCGUCGUGGGCCGCAAUUCAUUUUUCCCCUCACACA------ ((((((((((((((.....)))))))..((((-...(((((.((((.((-(........))).))))))))))))))))))))...........................------ ( -33.00, z-score = -2.76, R) >droSim1.chr2R 4896173 105 - 19596830 CCACGGCGAAUUCCUCAAAGGAAUUCGUGAAA-UUCGCAAAUGACUUCU-GGCAUUCCGUAGUGGUCUUUGCUU---UCGUGGGCCGCAUUUCAUUUUUCCCCUCGCAUA------ ((((((((((((((.....)))))))))....-...(((((.((((.((-(........))).)))))))))..---.)))))...........................------ ( -31.20, z-score = -2.24, R) >droSec1.super_1 3879814 105 - 14215200 CCACGGCGAAUUCCUCAAAGGAAUUCGUAAAA-UUCGCAAAUGACUUCU-GGCAUUCCGUAGUGGUCUUUGCUU---UCGUGGGCCGCAUUUCAUUUUUCCCCUCACAUA------ ((((((((((((((.....)))))))))....-...(((((.((((.((-(........))).)))))))))..---.)))))...........................------ ( -31.20, z-score = -3.18, R) >droYak2.chr2L 18898946 105 - 22324452 CCAAGGCGAAUUCCUCAAAGGAAUUUGUGAAA-UUCGCAAAUGACUUUU-CGCAUUCCGUAGUGGUCUUUGCUU---UCAUGGACCGCAUUUCAUCUUUUCCCUUACAUG------ ..((((.(((.........((((..(((((((-.(((....)))..)))-))))))))...(((((((.((...---.)).)))))))..........))))))).....------ ( -26.80, z-score = -2.45, R) >droEre2.scaffold_4929 18540194 104 + 26641161 CCGCGGCGAAUUCCUCUAAGGAAUUUGUGAAA-UUCGCAAAUGACUUUU-CGCAUUCCGUAGUGGUCUUUGCUU---UCAUGGCCCGCAUUUCAUC-UUUCCCUCACAUG------ ..((((((((((((.....)))))))((((((-...(((((.((((...-((.....))....)))))))))))---)))).).))))........-.............------ ( -26.50, z-score = -2.01, R) >droAna3.scaffold_13266 4734076 110 - 19884421 CCCUGGCAAAUUCCUUGCCGGAAUCUAGAAAAUUUUGCAAAUGACUUUA-CGCAUUCCGAAGUGGUCUUUGCUU---UCAUCGGCAUCGUUUCAUUUUCCCCCACCAGAACAUU-- ..(((((((.....)))))))......((((....(((..((((.....-.(((..((.....))....)))..---))))..)))...)))).....................-- ( -20.80, z-score = -0.13, R) >dp4.chr3 12604695 103 + 19779522 ------CCAAUCCUCCUUCGAUGGUACGGAA---UGGUAAAUGGCUUUUCCCCAUGCCGACAUGGUCUCUGCU----CUGUAGGCAUCAUUUCCCUUUCCCCCGCCCCCCCAUAUA ------................(((..((((---.((.(((((..........(((((.(((.(((....)))----.))).)))))))))).)).))))...))).......... ( -21.30, z-score = 0.06, R) >droPer1.super_2 1661958 103 + 9036312 ------CCAAUCCUCCUUCGAUGGUACGGAA---UGGUAAAUGGCUUUUCCCCAUGCCGACAUGGUCUCUGCU----CUGUAGGCAUCAUUUCCCUUUCCCCCGCCCCCCCAUAUA ------................(((..((((---.((.(((((..........(((((.(((.(((....)))----.))).)))))))))).)).))))...))).......... ( -21.30, z-score = 0.06, R) >consensus CCACGGCGAAUUCCUCAAAGGAAUUCGUGAAA_UUCGCAAAUGACUUUU_CGCAUUCCGUAGUGGUCUUUGCUU___UCGUGGGCCGCAUUUCAUUUUUCCCCUCACAUA______ .......(((((((.....)))))))..........(((((.((((.................)))))))))............................................ ( -9.95 = -10.36 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:17 2011