
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,455,830 – 2,455,944 |
| Length | 114 |
| Max. P | 0.719105 |

| Location | 2,455,830 – 2,455,944 |
|---|---|
| Length | 114 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Shannon entropy | 0.46833 |
| G+C content | 0.50521 |
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
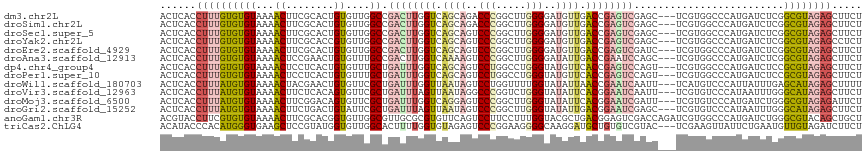
>dm3.chr2L 2455830 114 - 23011544 ACUCACCUUUGUGUGUAAAACUUCGCACUGUGUUGGCCGACUUGGUCAGCAGACCCGGCUUGGGGAUGUUGACCGAGUCGAGC---UCGUGGCCCAUGAUCUCGGCGUAGAGCUUCU .(((......(((((........))))).......((((((((((((((((..(((.....)))..)))))))))))))(((.---(((((...))))).))))))...)))..... ( -50.60, z-score = -3.66, R) >droSim1.chr2L 2416280 114 - 22036055 ACUCACCUUUGUGUGUAAAACUUCGCACUGUGUUGGCCGACUUGGUCAGCAGACCCGGCUUGGGGAUGUUGACCGAGUCGAGC---UCGUGGCCCAUGAUCUCGGCGUAGAGCUUCU .(((......(((((........))))).......((((((((((((((((..(((.....)))..)))))))))))))(((.---(((((...))))).))))))...)))..... ( -50.60, z-score = -3.66, R) >droSec1.super_5 629362 114 - 5866729 ACUCACCUUUGUGUGUAAAACUUCGCACUGUGUUGGCCGACUUGGUCAGCAGUCCCGGCUUGGGGAUGUUGACCGAGUCGAGC---UCGUGGCCCAUGAUCUCGGCGUAGAGCUUCU .(((......(((((........))))).......((((((((((((((((.((((......)))))))))))))))))(((.---(((((...))))).))))))...)))..... ( -49.90, z-score = -3.50, R) >droYak2.chr2L 2446086 114 - 22324452 ACUCACCUUUGUGUGUAAAACUUCGCACCGUGUUGGCCGACUUGGUCAGCAGUCCCGGCUUGGGGAUGUUGACCGAGUCGAGC---UCGUGGCCCAUGAUCUCGGCGUAGAGCCUCU .(((......(((((........))))).......((((((((((((((((.((((......)))))))))))))))))(((.---(((((...))))).))))))...)))..... ( -49.50, z-score = -3.16, R) >droEre2.scaffold_4929 2500003 114 - 26641161 ACUCACCUUUGUGUGUAAAACUUCGCACUGUGUUGGCCGACUUGGUCAGCAGUCCCGGCUUGGGGAUGUUGACCGAGUCGAUC---UCGUGGCCCAUGAUCUCGGCGUAGAGCUUCU .(((......(((((........))))).......((((((((((((((((.((((......)))))))))))))))))((..---(((((...)))))..)))))...)))..... ( -46.70, z-score = -2.95, R) >droAna3.scaffold_12913 196496 114 + 441482 ACUCACCUUUGUGUGUAAAACUCCGAACUGUGUUUGCCGACUUGGUCAAAAGUCCCGGCUUGGGGAUAUUGACCGAAUCCAGC---UCGUGGCCCAUGAUCUCGGCGUAGAGCUUCU ((.(((....))).))........(((((.((..((((((.((((((((..(((((......))))).)))))))).....(.---(((((...))))).))))))))).)).))). ( -32.10, z-score = -0.36, R) >dp4.chr4_group4 1721847 114 - 6586962 ACUCACCUUUGUGUGUAAAACUCCUCACUGUGUUUGCUGAUUUGGUCAGCAGUCCUGGCCUGGGUAUGUUCACCGAGUCCAGU---UCGUGGCCCAUGAUCUCCGCGUAGAGCUUCU ((.(((....))).))....(((..(........(((((((...)))))))(..((((.((.(((......))).)).)))).---.)((((..........)))))..)))..... ( -29.00, z-score = 0.25, R) >droPer1.super_10 725274 114 - 3432795 ACUCACCUUUGUGUGUAAAACUCCUCACUGUGUUUGCUGAUUUGGUCAGCAGUCCUGGCCUGGGUAUGUUCACCGAGUCCAGU---UCGUGGCCCAUGAUCUCCGCGUAGAGCUUCU ((.(((....))).))....(((..(........(((((((...)))))))(..((((.((.(((......))).)).)))).---.)((((..........)))))..)))..... ( -29.00, z-score = 0.25, R) >droWil1.scaffold_180703 2982946 114 - 3946847 ACUCACCUUUAUGUGUAAAACUACGAACUGUGUUCGCUGAUUUGGUUAAUAGUCCUGGUUUUGGUAUAUUAACCGAAUCAAUU---UCAUGUCCCAUUAUUUGAGCAUAGAGCUUUU ......((((((((.((((....(((((...))))).((((((((((((((..((.......))..))))))))))))))...---.............)))).))))))))..... ( -30.20, z-score = -3.43, R) >droVir3.scaffold_12963 9814147 114 - 20206255 ACUCACCUUUAUGUGUAAAACUUCUCACAGUGUUCGCUGAUUUAGUUAAUAGGCCCGGUCUGGGUAUAUUCACGGAAUCAAUU---UCGUGUCCCAUAAUUUGGGCAUAGAGCUUCU ......((((((......((((.....((((....))))....)))).....((((((..((((......(((((((....))---))))).))))....))))))))))))..... ( -28.50, z-score = -1.53, R) >droMoj3.scaffold_6500 30914969 114 + 32352404 ACUCACCUUUAUGUGUAAAACUUCGGACAGUGUUCGCUGAUUUGGUCAGGAGUCCCGGCUUGGGUAUAUUCACGGAAUCGAUU---UCGUGUCCCAUGAUCUGGGCGUAGAGAUUCU ......((((((((.....(((((.(((...(((....)))...))).))))).((((..((((......(((((((....))---))))).))))....))))))))))))..... ( -31.30, z-score = 0.05, R) >droGri2.scaffold_15252 5198239 114 + 17193109 ACUCACCUUUAUGUGUAAAACUUCUGACUGUAUUCGCUGAUUUAGUUAAUAGUCCCGGCUUGGGUAUAUUGACGGAAUCGAGC---UCGUGUCCCAUAAUUUGGGCAUAGAGCUUCU ............(((.((.((........)).))))).(((((.(((((((..(((.....)))..))))))).)))))((((---(((((.((((.....))))))).)))))).. ( -32.10, z-score = -2.04, R) >anoGam1.chr3R 49471208 117 + 53272125 ACGUACCUUCGUGUGUAAAACUUCGCACGGUGUUGGCGUUGCGCGUGUUCAGUCCUUCCUUUGGUACGCUGACGGAGUCGACCAGAUCGUGGCCCAUGAUCUGGGCGUACAGCUGCU ((((....(((((((........))))))).....)))).((((.....(((........)))((((((((((...)))).((((((((((...)))))))))))))))).)).)). ( -40.60, z-score = -0.34, R) >triCas2.ChLG4 12363227 114 - 13894384 ACAUACCCACAUGGGUGAAGCUCCGUAUGGUGUUGGCACUUUUGGUGUAGAGUCCCGGAAGGGGCAAGGAUGCUGUGUCGUAC---UCGAAGUUAUUCUGAAUGUUGUAGAUCUUCU ...(((((....)))))..((.(((...((((....))))..))).))...(((((....)))))..(..(((......))).---.)((((....((((.......)))).)))). ( -33.90, z-score = -0.46, R) >consensus ACUCACCUUUGUGUGUAAAACUUCGCACUGUGUUGGCCGAUUUGGUCAGCAGUCCCGGCUUGGGUAUGUUGACCGAGUCGAGC___UCGUGGCCCAUGAUCUCGGCGUAGAGCUUCU ......((((((((((...((........))....)).((((((((.((((..(((.....)))..)))).)))))))).........................))))))))..... (-18.44 = -18.59 + 0.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:23 2011