| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,260,523 – 6,260,648 |
| Length | 125 |
| Max. P | 0.995327 |

| Location | 6,260,523 – 6,260,648 |
|---|---|
| Length | 125 |
| Sequences | 8 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 71.25 |
| Shannon entropy | 0.54871 |
| G+C content | 0.41315 |
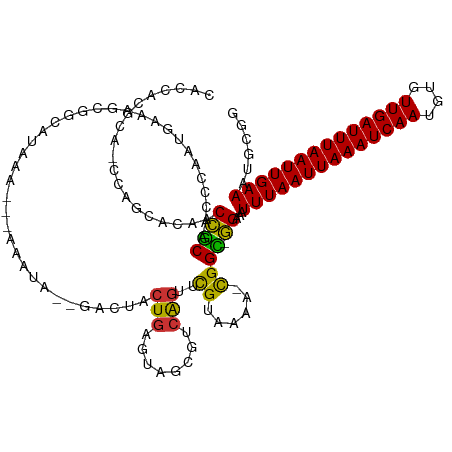
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.66 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
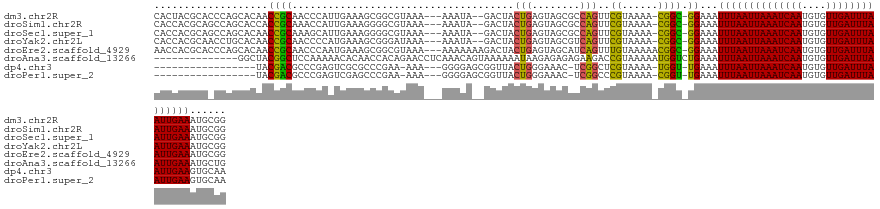
>dm3.chr2R 6260523 125 + 21146708 CACUACGCACCCAGCACAACCGCAACCCAUUGAAAGCGGCGUAAA---AAAUA--GACUACUGAGUAGCGCCAGUUCGUAAAA-CGGC-GGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCGG .....((((..........((((.......((((...(((((...---...((--(....)))....)))))..)))).....-..))-))...((((((((((((((....)))))))))))))).)))). ( -33.74, z-score = -2.58, R) >droSim1.chr2R 4883440 125 + 19596830 CACCACGCAGCCAGCACCACCGCAAACCAUUGAAAGGGGCGUAAA---AAAUA--GACUACUGAGUAGCGCCAGUUCGUAAAA-CGGC-GGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCGG .....((((..........((((.......((((...(((((...---...((--(....)))....)))))..)))).....-..))-))...((((((((((((((....)))))))))))))).)))). ( -35.14, z-score = -2.53, R) >droSec1.super_1 3867240 125 + 14215200 CACCACGCAGCCAGCACAACCGCAAAGCAUUGAAAGGGGCGUAAA---AAAUA--GACUACUGAGUAGCGCCAGUUCGUAAAA-CGGC-GGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCGG .....((((..........((((.......((((...(((((...---...((--(....)))....)))))..)))).....-..))-))...((((((((((((((....)))))))))))))).)))). ( -35.14, z-score = -2.54, R) >droYak2.chr2L 18887317 125 + 22324452 CACCACGCAACCUGCACAACCGCAACCCCAUGAAAGCGGGAUAAA---AAAUA--GACUACUGAGUAGCGUCAGUUCGUAAAA-CGGC-GGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCGG .....((((..........((((...(((.(....).))).....---.....--((..(((((......)))))))......-..))-))...((((((((((((((....)))))))))))))).)))). ( -33.00, z-score = -2.64, R) >droEre2.scaffold_4929 18529161 128 - 26641161 AACCACGCACCCAGCACAACCGCAACCCAAUGAAAGCGGCGUAAA---AAAAAAAGACUACUGAGUAGCAUCAGUUUGUAAAAACGGC-GGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCGG .....((((.((.((....((((............))))......---........((.(((((......)))))..)).......))-))...((((((((((((((....)))))))))))))).)))). ( -30.60, z-score = -2.20, R) >droAna3.scaffold_13266 4723644 118 + 19884421 --------------GGCUACGGCUCCAAAAACACAACCACAGAACCUCAAACAGUAAAAAAUAAGAGAGAGAAGACCGUAAAAAUGGUCUGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCUG --------------(((....)))...........................(((((................(((((((....)))))))....((((((((((((((....)))))))))))))).))))) ( -27.50, z-score = -3.82, R) >dp4.chr3 12593930 108 - 19779522 -----------------UACGACGCCCGAGUCGCGCCCGAA-AAA---GGGGAGCGGUUACUGGGAAAC-UCGGCUCGUAAAA-UGGU-UGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAGUGCAA -----------------(((((.(((..(..(((.(((...-...---)))..)))..)...((....)-).))))))))...-...(-((...((((((((((((((....))))))))))))))...))) ( -35.80, z-score = -3.16, R) >droPer1.super_2 1651197 108 - 9036312 -----------------UACGACGCCCGAGUCGAGCCCGAA-AAA---GGGGAGCGGUUACUGGGAAAC-UCGGCCCGUAAAA-CGGU-UGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAGUGCAA -----------------..(((..(((.(..((..(((...-...---.)))..))..)...)))....-)))..(((.....-)))(-((...((((((((((((((....))))))))))))))...))) ( -32.20, z-score = -1.94, R) >consensus CACCACGCA_CCAGCACAACCGCAACCCAAUGAAAGCGGCAUAAA___AAAUA__GACUACUGAGUAGCGUCAGUUCGUAAAA_CGGC_GGAAAUUUAAUUAAAUCAAUGUGUUGAUUUAAUUGAAAUGCGG ...................(((((....................................(((........)))....................((((((((((((((....)))))))))))))).))))) (-15.04 = -15.66 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:15 2011