
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,190,970 – 6,191,094 |
| Length | 124 |
| Max. P | 0.976303 |

| Location | 6,190,970 – 6,191,086 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Shannon entropy | 0.17287 |
| G+C content | 0.31964 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.86 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6190970 116 - 21146708 AAAAUCACAGUGCUAUCAAU-GUGAUAGUGUUAUCUUCUU--UUAUAAACAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUAGGCAGCCUGUGUUAUAUUAAUACGUUAU ........((.((((((...-.(((((((...((((..((--....))..))))...))))))).)))))).))...(((((((((..((....))..)))))))))............ ( -26.50, z-score = -1.95, R) >droEre2.scaffold_4929 18457149 116 + 26641161 AAAAUCGCAGUGCUAUCAAUGGUGAUAGUGUUAUCUUCUU--UUAUAAG-AGGUUUAACUAUCAAGGUUUAACUAUUUGUAGCACAUUCUCGGCAGCAUGUGCUAUAUUAACACGCUAU ......((.(((.....(((..(((((((.....((((((--....)))-)))....)))))))..)))........((((((((((.((....)).))))))))))....)))))... ( -30.60, z-score = -2.82, R) >droYak2.chr2L 18819096 118 - 22324452 AAAAUCGCAGUGCUAUCAAU-GUGAUAGUGUUAUCUUUUCAGUUAUACAGAGGUUUAACUAUCAAGGUGAUUCUAUUUGUAGCACUUUCUUGGUAGCAUGUGUUAUAUUAUUAACUUAU .....((((.(((((((((.-..((((((...((((((..........))))))...))))))((((((...(.....)...)))))).)))))))))))))................. ( -28.10, z-score = -2.13, R) >droSec1.super_1 3798406 116 - 14215200 AAAAUCGCAGUGCUAUCAAU-GUGAUAGUGUUAUCUUCUU--UUAUAAAGAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUUGACAGCAUGUGUUAUAUUAAUACGUUAU ........((.((((((...-.(((((((.....((((((--.....))))))....))))))).)))))).))...((((((((((.((....)).))))))))))............ ( -32.00, z-score = -3.61, R) >droSim1.chr2R 4814269 116 - 19596830 AAAAUCGCAGUGCUAUCAAU-GUGAUAGUGUUAUCUUCUU--UUAUAAAGAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUUGGCAGCAUGUGUUAUAUUAAUACGUUAU ........((.((((((...-.(((((((.....((((((--.....))))))....))))))).)))))).))...((((((((((.((....)).))))))))))............ ( -32.00, z-score = -3.48, R) >consensus AAAAUCGCAGUGCUAUCAAU_GUGAUAGUGUUAUCUUCUU__UUAUAAAGAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUUGGCAGCAUGUGUUAUAUUAAUACGUUAU ........((.((((((.....(((((((.....((((((.......))))))....))))))).)))))).))...((((((((((.((....)).))))))))))............ (-23.70 = -24.86 + 1.16)

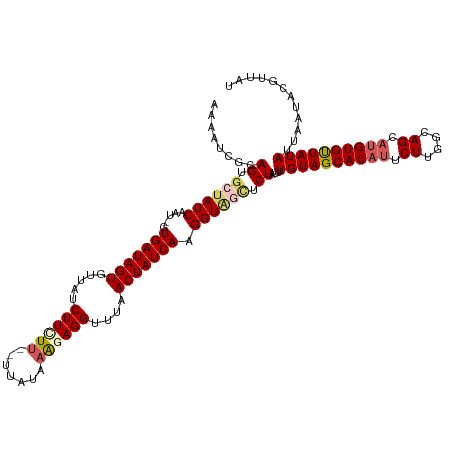
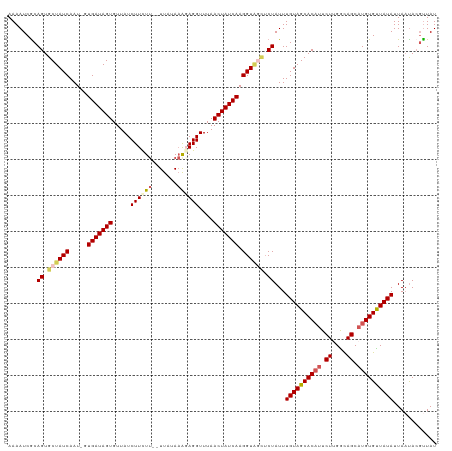
| Location | 6,190,980 – 6,191,094 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Shannon entropy | 0.15327 |
| G+C content | 0.34619 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.86 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6190980 114 - 21146708 CACGGAGAAAAAUCACAGUGCUAUCAAU-GUGAUAGUGUUAUCUUCUU--UUAUAAACAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUAGGCAGCCUGUGUUAUAUU ................((.((((((...-.(((((((...((((..((--....))..))))...))))))).)))))).))...(((((((((..((....))..))))))))).. ( -26.50, z-score = -1.39, R) >droEre2.scaffold_4929 18457159 114 + 26641161 CACAGAGAAAAAUCGCAGUGCUAUCAAUGGUGAUAGUGUUAUCUUCUU--UUAUAAG-AGGUUUAACUAUCAAGGUUUAACUAUUUGUAGCACAUUCUCGGCAGCAUGUGCUAUAUU ....(((((........(((((((.((((((((((((.....((((((--....)))-)))....))))))........)))))).))))))).)))))((((.....))))..... ( -29.80, z-score = -2.48, R) >droYak2.chr2L 18819106 116 - 22324452 CACAGAGAAAAAUCGCAGUGCUAUCAAU-GUGAUAGUGUUAUCUUUUCAGUUAUACAGAGGUUUAACUAUCAAGGUGAUUCUAUUUGUAGCACUUUCUUGGUAGCAUGUGUUAUAUU .............((((.(((((((((.-..((((((...((((((..........))))))...))))))((((((...(.....)...)))))).)))))))))))))....... ( -28.10, z-score = -1.56, R) >droSec1.super_1 3798416 114 - 14215200 CACGGAGAAAAAUCGCAGUGCUAUCAAU-GUGAUAGUGUUAUCUUCUU--UUAUAAAGAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUUGACAGCAUGUGUUAUAUU ................((.((((((...-.(((((((.....((((((--.....))))))....))))))).)))))).))...((((((((((.((....)).)))))))))).. ( -32.00, z-score = -3.07, R) >droSim1.chr2R 4814279 114 - 19596830 CACGGAGAAAAAUCGCAGUGCUAUCAAU-GUGAUAGUGUUAUCUUCUU--UUAUAAAGAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUUGGCAGCAUGUGUUAUAUU ................((.((((((...-.(((((((.....((((((--.....))))))....))))))).)))))).))...((((((((((.((....)).)))))))))).. ( -32.00, z-score = -2.97, R) >consensus CACGGAGAAAAAUCGCAGUGCUAUCAAU_GUGAUAGUGUUAUCUUCUU__UUAUAAAGAGGUUUAACUAUCAAGGUAGCUCUAUUUGUAGCACAUUCUUGGCAGCAUGUGUUAUAUU ................((.((((((.....(((((((.....((((((.......))))))....))))))).)))))).))...((((((((((.((....)).)))))))))).. (-23.70 = -24.86 + 1.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:12 2011