| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,178,732 – 6,178,824 |
| Length | 92 |
| Max. P | 0.631661 |

| Location | 6,178,732 – 6,178,824 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Shannon entropy | 0.45602 |
| G+C content | 0.41342 |
| Mean single sequence MFE | -16.74 |
| Consensus MFE | -11.55 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6178732 92 + 21146708 CUUUUGAUACGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAACACCAACAUCAGCACCACCUCA--------- ...(((((..((((---(.......)))))..((((((((....-.......)))))))).....................)))))..........--------- ( -14.50, z-score = -0.91, R) >droSim1.chr2R 4801234 92 + 19596830 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAACACCAGCAUCAGCACCACCUCA--------- ....((.(((((((---(.......))))...((((((((....-.......)))))))).........)))).))...((....)).........--------- ( -15.50, z-score = -0.68, R) >droSec1.super_1 3786105 92 + 14215200 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAACACCAGCAUCAGCACCACCUCA--------- ....((.(((((((---(.......))))...((((((((....-.......)))))))).........)))).))...((....)).........--------- ( -15.50, z-score = -0.68, R) >droYak2.chr2L 18806747 91 + 22324452 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAUCACCAGCAUCAGCACACCCCA---------- ....((((((((((---(.......))))...((((((((....-.......)))))))).........)))))))...((....))........---------- ( -20.10, z-score = -2.59, R) >droEre2.scaffold_4929 18444701 84 - 26641161 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAACACCAGCAUCAGCA----------------- ....((.(((((((---(.......))))...((((((((....-.......)))))))).........)))).))...((....)).----------------- ( -15.50, z-score = -0.62, R) >droAna3.scaffold_13266 4642410 98 + 19884421 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGACCGACCAGCAUCACCACCCCCACUGCUCCA--- ........(((.((---((((...........((((((((....-.......))))))))(....))))))).)))..((((.............))))...--- ( -16.32, z-score = -1.14, R) >dp4.chr3 12510430 84 - 19779522 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAACAUCAG-ACCUCCGC---------------- ..((((((((((((---(.......))))...((((((((....-.......)))))))).........)))...)))))-).......---------------- ( -15.70, z-score = -0.93, R) >droPer1.super_2 1566779 84 - 9036312 CUUUUGAUUCGAUU---GUAUAAUUCAAUUAAGUGCUGAUUGAA-UAAAGACGUCAGCGCGCCCACAUACGAAACAUCAG-ACCUCCGC---------------- ..((((((((((((---(.......))))...((((((((....-.......)))))))).........)))...)))))-).......---------------- ( -15.70, z-score = -0.93, R) >droWil1.scaffold_181009 959214 104 - 3585778 UUAUAAAUUCCAUUUUUGUAUAAUUCAAUUAAGUGCUGAUUGAAAUAAAGACGUCAGCGCGCCCACAUUCAAUAUUAUAGGAAAAGC-UGUUGAAAAUCUGCACA .((((((.......))))))............((((((((............))))))))((.....(((((((.............-))))))).....))... ( -16.02, z-score = 0.06, R) >droMoj3.scaffold_6496 19166616 100 - 26866924 -CUUUGAUACGAUU---GUAUAAUUCGAUCAAGUGCUGAUUGAA-UUAAGACGUCAGCGCAUCCACCCACUAUAAAAAUACAGCAGAGUGAGGCAGAGAUGGGGA -..(((((.(((..---.......))))))))((((((((....-.......)))))))).....((((((.......(((......)))......)).)))).. ( -22.52, z-score = -0.92, R) >consensus CUUUUGAUUCGAUU___GUAUAAUUCAAUUAAGUGCUGAUUGAA_UAAAGACGUCAGCGCGCCCACAUACGAAACACCAGCAUCAGCACCACC_C__________ ...((((((.((((.......)))).))))))((((((((............))))))))............................................. (-11.55 = -11.77 + 0.22)

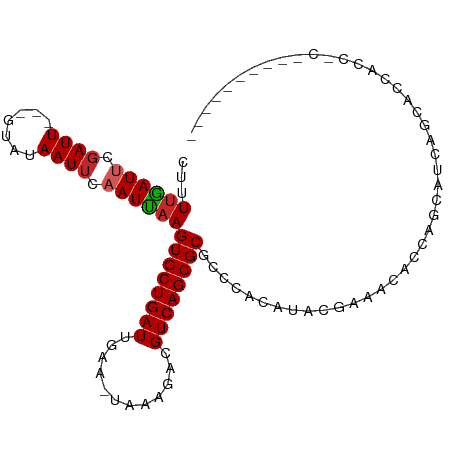

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:10 2011