| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,152,348 – 6,152,432 |
| Length | 84 |
| Max. P | 0.593228 |

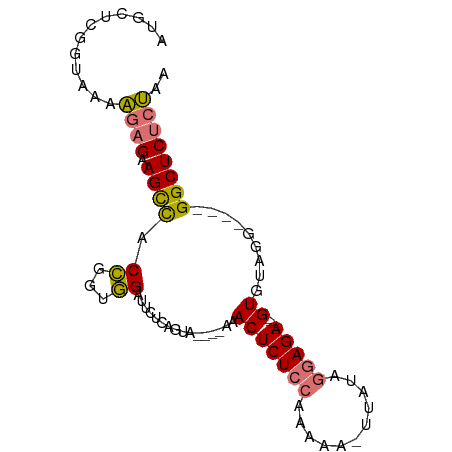
| Location | 6,152,348 – 6,152,432 |
|---|---|
| Length | 84 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.44960 |
| G+C content | 0.44151 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.72 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.593228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
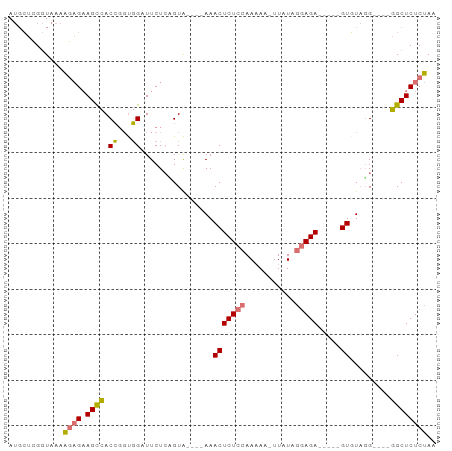
>dm3.chr2R 6152348 84 + 21146708 AUGCUCGGUAAAAGAGAAGCCACCGGCGGAUUCUCAGUG----AAACUCUCCAAUAA-UUAUAGGAGA-----GUGUAUGUUCUGGCUCUCUAA ............(((((.((((..((((..(((.....)----))(((((((.....-.....)))))-----))...)))).))))))))).. ( -23.30, z-score = -1.41, R) >droAna3.scaffold_13266 4616001 91 + 19884421 ---AGAGAUAGAGUCGCAGUUACACCUUGAUUCUCCGUAUAGGAAACUCUCCAAAAAACUAUCAAAGAAAUCGGUUUCGGCGAGGGCUCUCCAC ---.((((..((((((.((......))))))))(((.....)))..((((((...(((((............)))))..).))))).))))... ( -16.90, z-score = 0.74, R) >droSec1.super_1 3758755 80 + 14215200 AUGCUCGGUAAAAGAGAAGCCACCGGUGGUUUCUAGGUA----AAACUCUCCAAAAA-UUAUAGGAGA-----GUGUAGG----GGCUCUCUAA ..((((........((((((((....)))))))).....----..(((((((.....-.....)))))-----))....)----)))....... ( -25.70, z-score = -2.51, R) >droSim1.chr2R 4776574 80 + 19596830 AUGCUCGGUAAAAGAGAAGCCACCGGUGGUUUCUCUGUA----AAACUCUCCAAAAA-UUAUAGGAGA-----GUGUAGG----GGCUCUCUAA ..((((......((((((((((....))))))))))...----..(((((((.....-.....)))))-----))....)----)))....... ( -30.10, z-score = -4.17, R) >consensus AUGCUCGGUAAAAGAGAAGCCACCGGUGGAUUCUCAGUA____AAACUCUCCAAAAA_UUAUAGGAGA_____GUGUAGG____GGCUCUCUAA ............((((.((((.((...))..................(((((...........)))))................)))))))).. (-10.85 = -10.72 + -0.13)

| Location | 6,152,348 – 6,152,432 |
|---|---|
| Length | 84 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.44960 |
| G+C content | 0.44151 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -9.51 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
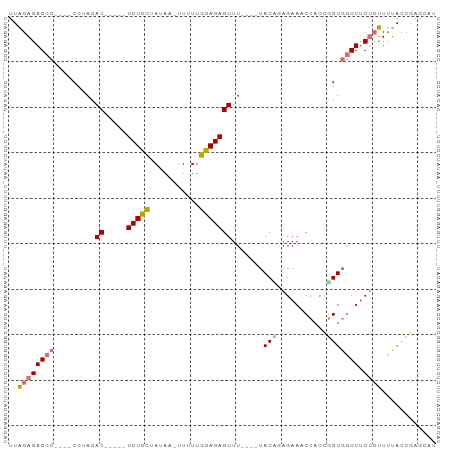
>dm3.chr2R 6152348 84 - 21146708 UUAGAGAGCCAGAACAUACAC-----UCUCCUAUAA-UUAUUGGAGAGUUU----CACUGAGAAUCCGCCGGUGGCUUCUCUUUUACCGAGCAU ..((((((((.(((.....((-----(((((.....-.....)))))))))----)((((.(......)))))))).)))))............ ( -22.70, z-score = -2.08, R) >droAna3.scaffold_13266 4616001 91 - 19884421 GUGGAGAGCCCUCGCCGAAACCGAUUUCUUUGAUAGUUUUUUGGAGAGUUUCCUAUACGGAGAAUCAAGGUGUAACUGCGACUCUAUCUCU--- ..(((((....((((.(..(((...(((((..(.......)..)))))(((((.....))))).....)))....).)))).....)))))--- ( -22.20, z-score = -0.16, R) >droSec1.super_1 3758755 80 - 14215200 UUAGAGAGCC----CCUACAC-----UCUCCUAUAA-UUUUUGGAGAGUUU----UACCUAGAAACCACCGGUGGCUUCUCUUUUACCGAGCAU ..((((((((----((...((-----(((((.....-.....)))))))..----......(....)...)).))).)))))............ ( -21.20, z-score = -2.65, R) >droSim1.chr2R 4776574 80 - 19596830 UUAGAGAGCC----CCUACAC-----UCUCCUAUAA-UUUUUGGAGAGUUU----UACAGAGAAACCACCGGUGGCUUCUCUUUUACCGAGCAU .......((.----.....((-----(((((.....-.....)))))))..----...((((((.(((....))).))))))........)).. ( -21.60, z-score = -2.61, R) >consensus UUAGAGAGCC____CCUACAC_____UCUCCUAUAA_UUUUUGGAGAGUUU____UACAGAGAAACCACCGGUGGCUUCUCUUUUACCGAGCAU ..((((((((................(((((...........))))).........((((.........)))))))).))))............ ( -9.51 = -10.20 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:06 2011