
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,104,826 – 6,104,960 |
| Length | 134 |
| Max. P | 0.878034 |

| Location | 6,104,826 – 6,104,926 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 97.56 |
| Shannon entropy | 0.04332 |
| G+C content | 0.38115 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
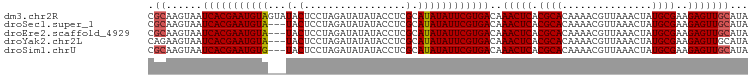
>dm3.chr2R 6104826 100 + 21146708 CGCAAGUAAUCACGAAUGUAGUAUACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUA .((......((((((((((((((((..........)))))......)))))))))))..(((((.((((...............))))..)))))))... ( -21.46, z-score = -1.75, R) >droSec1.super_1 3711360 97 + 14215200 CGCAAGUAAUCACGAAUGUA---UACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUA .((......(((((((((((---(.(.................).))))))))))))..(((((.((((...............))))..)))))))... ( -21.19, z-score = -2.24, R) >droEre2.scaffold_4929 18367762 97 - 26641161 CGCAAGUAAUCACGAAUGUA---UACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUA .((......(((((((((((---(.(.................).))))))))))))..(((((.((((...............))))..)))))))... ( -21.19, z-score = -2.24, R) >droYak2.chr2L 18730553 97 + 22324452 CAGAAGUAAUCACGAAUGUA---UACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUA .....((..(((((((((((---(.(.................).))))))))))))..(((((.((((...............))))..)))))))... ( -18.89, z-score = -1.64, R) >droSim1.chrU 6974573 97 - 15797150 CGCAAGUAAUCACGAAUGUG---UACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUA .((......(((((((((((---(.(.................).))))))))))))..(((((.((((...............))))..)))))))... ( -21.19, z-score = -1.72, R) >consensus CGCAAGUAAUCACGAAUGUA___UACUCCUAGAUAUAUACCUCGCAUAUAUUCGUGACAAACUCACGCACAAAACGUUAAACUAUGCGAAGAGUUGCAUA .((......(((((((((((.........((....)).........)))))))))))..(((((.((((...............))))..)))))))... (-17.99 = -18.23 + 0.24)


| Location | 6,104,826 – 6,104,926 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 97.56 |
| Shannon entropy | 0.04332 |
| G+C content | 0.38115 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6104826 100 - 21146708 UAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUAUACUACAUUCGUGAUUACUUGCG ...(((((((..(((((((.........))))))).))))..(((((((((.(((((.((((....))))....)))))....)))))))))....))). ( -28.10, z-score = -1.86, R) >droSec1.super_1 3711360 97 - 14215200 UAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUA---UACAUUCGUGAUUACUUGCG ...(((((((..(((((((.........))))))).))))..(((((((((.(((((.((((....))))....)))---)).)))))))))....))). ( -31.10, z-score = -3.14, R) >droEre2.scaffold_4929 18367762 97 + 26641161 UAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUA---UACAUUCGUGAUUACUUGCG ...(((((((..(((((((.........))))))).))))..(((((((((.(((((.((((....))))....)))---)).)))))))))....))). ( -31.10, z-score = -3.14, R) >droYak2.chr2L 18730553 97 - 22324452 UAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUA---UACAUUCGUGAUUACUUCUG .....(((((..(((((((.........))))))).))))).(((((((((.(((((.((((....))))....)))---)).)))))))))........ ( -28.90, z-score = -2.82, R) >droSim1.chrU 6974573 97 + 15797150 UAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUA---CACAUUCGUGAUUACUUGCG ...(((((((..(((((((.........))))))).))))..(((((((((.(.(((.((((....))))....)))---.).)))))))))....))). ( -27.00, z-score = -1.64, R) >consensus UAUGCAACUCUUCGCAUAGUUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAGGUAUAUAUCUAGGAGUA___UACAUUCGUGAUUACUUGCG ...(((((((..(((((((.........))))))).))))..(((((((((...(((.((((....))))....)))......)))))))))....))). (-24.26 = -24.30 + 0.04)



| Location | 6,104,866 – 6,104,960 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.47546 |
| G+C content | 0.45864 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -11.57 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6104866 94 - 21146708 CAGUCCCAAUCCCCAAC-CAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAG .................-....(((((((.((......(((((((..(((((((--.........))))))).))).))))....)).))))))).. ( -18.60, z-score = -1.09, R) >droSec1.super_1 3711397 89 - 14215200 ------CAAUUCCCCUAACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAG ------................(((((((.((......(((((((..(((((((--.........))))))).))).))))....)).))))))).. ( -18.60, z-score = -1.06, R) >droEre2.scaffold_4929 18367799 89 + 26641161 ------CAUUCCCCACCACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAG ------..........(((((..((((.......((((((........))))))--....)))).)))))(((((.....)))))............ ( -18.80, z-score = -1.05, R) >droYak2.chr2L 18730590 95 - 22324452 CGCUCCCUCAUUCCCCCACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAG (((.............(((((..((((.......((((((........))))))--....)))).)))))(((((.....))))).......))).. ( -22.30, z-score = -2.30, R) >droAna3.scaffold_13266 4572523 87 - 19884421 --------AAAUGCCACACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAACGUUUUGUGCGUGAGUCUGUCCCACCACACCACCAC --------.........((((.((((((.......))))))((((..(((((((--.........))))))).))))))))................ ( -17.60, z-score = -1.36, R) >dp4.chr3 6940176 85 - 19779522 --------CACUCGGGCGCAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAGCGUUUUGUGCGUGCGCUUGUCGCACAUGAAUACC-- --------.....((((((((..((((.......((((((........))))))--....)))).))))))(((((.....))))).......))-- ( -25.90, z-score = -1.04, R) >droPer1.super_2 7142384 85 - 9036312 --------CACUCGGGCGCAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAGCGUUUUGUGCGUGCGCUUGUCGCACAUGAAUACC-- --------.....((((((((..((((.......((((((........))))))--....)))).))))))(((((.....))))).......))-- ( -25.90, z-score = -1.04, R) >droMoj3.scaffold_6496 19062152 86 + 26866924 ---------CAGCAGCUGCUGCUGCGUAUCUUAUCUAUGCAACUC-UCGCAUAGUAUUUAAUG-UGCGUGAGCUACACAGCCAGACACACACACAUA ---------..(((((....)))))((.(((...((.((...(((-.((((((........))-)))).)))...)).))..))).))......... ( -24.40, z-score = -0.99, R) >droSim1.chrU 6974610 89 + 15797150 ------CAAUUCCCCUAACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG--UUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAG ------................(((((((.((......(((((((..(((((((--.........))))))).))).))))....)).))))))).. ( -18.60, z-score = -1.06, R) >consensus ______C_AACCCCCCCACAGCUGCGUAUCUUAUCUAUGCAACUCUUCGCAUAG__UUUAACGUUUUGUGCGUGAGUUUGUCACGAAUAUAUGCGAG ................(((((..((((.......((((((........))))))......)))).))))).(((.......)))............. (-11.57 = -12.03 + 0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:12:01 2011