| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,086,141 – 6,086,246 |
| Length | 105 |
| Max. P | 0.592490 |

| Location | 6,086,141 – 6,086,246 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.77 |
| Shannon entropy | 0.65594 |
| G+C content | 0.51676 |
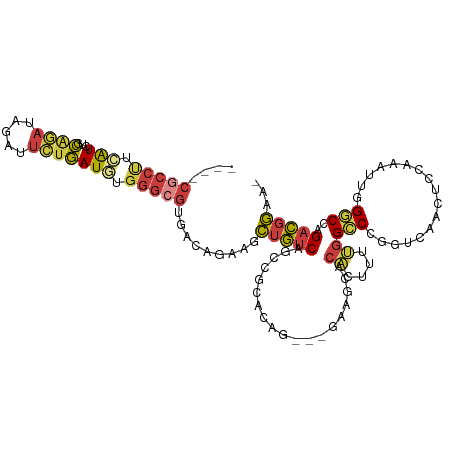
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -12.56 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.592490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
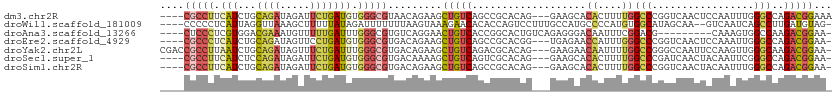
>dm3.chr2R 6086141 105 + 21146708 ----CGCCUUCAUCUGCAGAUAGAUUCUGAUGUGGGCGUAACAGAAGCUGUCAGCCGCACAG---GAAGCACACUUUUGGCCCGGUCAACUCCAAUUUGGGCCAGACGGAAA ----.((.(((.....((((.....)))).(((((((...((((...))))..))).)))))---)).))...(.((((((((((...........)))))))))).).... ( -34.20, z-score = -1.00, R) >droWil1.scaffold_181009 821059 105 - 3585778 ----CCCCCUCAUUAGGUUAAAGCUUUUUAUAGAUUUUUUUAAGUAAAGAACACACCAGUCCUUUGCCAUGCCCCAUGUGGCAUAGCAA--GUCAAUCAGCCUUGAUGUAG- ----....(((((((((((....((((...((((....))))...)))).............(((((.(((((......))))).))))--)......)))).))))).))- ( -19.70, z-score = -1.15, R) >droAna3.scaffold_13266 4551955 98 + 19884421 ----CUCCCUCGUGGACGAAAUGUUUUUGAUUUGGGCGUGUCAGGAACUGUCACCGGCACUGUCAGAGGGACAAUUUCGGACG---------CAAAGUGGCCAAGACGGAA- ----.(((.(((((..(((((((((((((((...(.((((.(((...))).)).)).)...))))))..))).))))))..))---------)...(....)..)).))).- ( -27.30, z-score = 0.76, R) >droEre2.scaffold_4929 18348808 104 - 26641161 ----CGCCCUCAUCUGCAGAUAGUUCCUGAUGUGGGCGUGACAGAAGCUGUCAGCCGCACGG---UGAGAACCAUUUGGGCCCGGUCAACUCCAAAUUGGGCCAGACGGAA- ----..((.((.....(((((.(((((...(((((((.((((((...))))))))).)))).---.).)))).)))))((((((((.........)))))))).)).))..- ( -40.00, z-score = -1.58, R) >droYak2.chr2L 18711751 108 + 22324452 CGACCGCCUUAAUCUGCAGAUAGUUUCUGAUUUGGGCGUGACAGAAGCUGUCAGACGCACAG---GAAGAACAAUUUUGGCCGGGCCAAUUCCAAGUUGGGCAAGACGGAA- ...(((.(((.(((....)))....((..(((((((((((((((...)))))..))))....---...........(((((...)))))..))))))..)).))).)))..- ( -30.00, z-score = -0.08, R) >droSec1.super_1 3692598 104 + 14215200 ----CGCCUUCAUCUCCAGAUAGAUUCUGAUGUGGGCGUGACAAAAGCUGUCAGUCGCACAG---GAAGCACACUUUUGGCCCGAUCAACUACAAUUCGGGCCAGACGGAA- ----.((.(((.....((((.....)))).(((((((.(((((.....)))))))).)))))---)).))...(.((((((((((...........)))))))))).)...- ( -37.70, z-score = -2.94, R) >droSim1.chr2R 4708766 104 + 19596830 ----CGCCUUCAUCUGCAGAUAGAUUCUGAUGUGGGCGUGACAGAAGCUGUCAGCCGCACAG---GAAGCACACUUUUGGCCCGGUCAACUACAAUUUGGGCCAGACGGAA- ----.((.(((.....((((.....)))).(((((((.((((((...))))))))).)))))---)).))...(.((((((((((...........)))))))))).)...- ( -38.90, z-score = -2.36, R) >consensus ____CGCCUUCAUCUGCAGAUAGAUUCUGAUGUGGGCGUGACAGAAGCUGUCAGCCGCACAG___GAAGCACACUUUUGGCCCGGUCAACUCCAAAUUGGGCCAGACGGAA_ ....(((((.(((...((((.....))))))).))))).........(((((...................((....))(((.................)))..)))))... (-12.56 = -12.83 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:58 2011