
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 6,053,581 – 6,053,674 |
| Length | 93 |
| Max. P | 0.896664 |

| Location | 6,053,581 – 6,053,674 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.37078 |
| G+C content | 0.56750 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6053581 93 + 21146708 -----------CCCUUCGUGGCCGUGGUCCG--AGGACUCCCAAGUCCC------CGGAUCCUAAGUCCUAAACACCGUGUACGUGCGGACACGCAAAAAUUGUUCACCAAU -----------.....((((.((((((((((--.(((((....))))).------))))))...............((....)).)))).)))).................. ( -26.50, z-score = -1.34, R) >droSim1.chr2R 4683920 90 + 19596830 --------------CCCUUUGCCGUGGUCCG--AGGACUCCCAAGUCCC------CGGAUCCUAAGUCCUAAACACCGUGUACGUGCGGACACGCAAAAAUUGUUCACCAAU --------------...(((((.((((((((--.(((((....))))).------)))))...............((((......)))).)))))))).............. ( -26.70, z-score = -2.46, R) >droSec1.super_1 3669322 90 + 14215200 --------------CCCUUUGCCGUGGUCCG--AGGACUCCCAAGUCCC------CGGAUCCUAAGUCCUAAACACCGUGUACGUGCGGACACGCAAAAAUUGUUCACCAAU --------------...(((((.((((((((--.(((((....))))).------)))))...............((((......)))).)))))))).............. ( -26.70, z-score = -2.46, R) >droYak2.chr2L 18687837 90 + 22324452 --------------CCCUUUGCAGUGGUCCG--AGGACUCCCAAGUCCC------CGGAUCCUAAGUCCCGAACACCGUGUACGUGCGGACACGCAAAAAUUGUUCACCAAU --------------...(((((((.((((((--.(((((....))))).------))))))))..((((((.((.....)).))...))))..))))).............. ( -28.80, z-score = -2.74, R) >droPer1.super_2 7102767 112 + 9036312 UGGUCCUGCCCCUCCUCCUUCUCGAGUCCAGUCGCGAGUCCCGAGUCCCGUGCUCCGAGUCCCGACUCCCGAGUCUCGUGUACGUGCGGACACGCAAAAAUUGCAGAGCCCU .((..((((.....(((......))).......(((.((((((.....(((((..((((.(.((.....)).).)))).))))))).)))).))).......))))..)).. ( -34.50, z-score = -1.65, R) >consensus ______________CCCUUUGCCGUGGUCCG__AGGACUCCCAAGUCCC______CGGAUCCUAAGUCCUAAACACCGUGUACGUGCGGACACGCAAAAAUUGUUCACCAAU ..................((((.(((.......((((((.....((((........))))....)))))).....((((......)))).)))))))............... (-15.86 = -16.02 + 0.16)

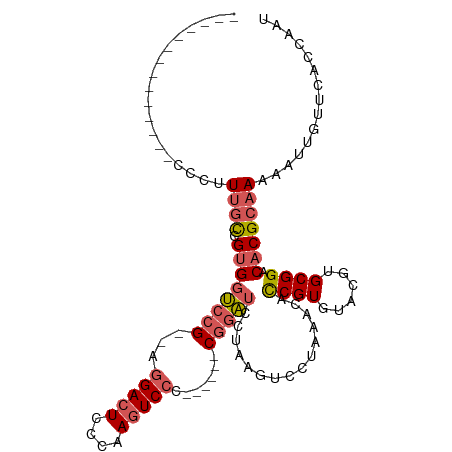

| Location | 6,053,581 – 6,053,674 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Shannon entropy | 0.37078 |
| G+C content | 0.56750 |
| Mean single sequence MFE | -36.16 |
| Consensus MFE | -24.16 |
| Energy contribution | -22.92 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 6053581 93 - 21146708 AUUGGUGAACAAUUUUUGCGUGUCCGCACGUACACGGUGUUUAGGACUUAGGAUCCG------GGGACUUGGGAGUCCU--CGGACCACGGCCACGAAGGG----------- ..(((((((((.....((((((....)))))).....)))))........((.((((------((((((....))))))--))))))...))))(....).----------- ( -35.50, z-score = -2.01, R) >droSim1.chr2R 4683920 90 - 19596830 AUUGGUGAACAAUUUUUGCGUGUCCGCACGUACACGGUGUUUAGGACUUAGGAUCCG------GGGACUUGGGAGUCCU--CGGACCACGGCAAAGGG-------------- .(((.....)))(((((((..((((((((.......))))...))))...((.((((------((((((....))))))--))))))...))))))).-------------- ( -34.40, z-score = -2.42, R) >droSec1.super_1 3669322 90 - 14215200 AUUGGUGAACAAUUUUUGCGUGUCCGCACGUACACGGUGUUUAGGACUUAGGAUCCG------GGGACUUGGGAGUCCU--CGGACCACGGCAAAGGG-------------- .(((.....)))(((((((..((((((((.......))))...))))...((.((((------((((((....))))))--))))))...))))))).-------------- ( -34.40, z-score = -2.42, R) >droYak2.chr2L 18687837 90 - 22324452 AUUGGUGAACAAUUUUUGCGUGUCCGCACGUACACGGUGUUCGGGACUUAGGAUCCG------GGGACUUGGGAGUCCU--CGGACCACUGCAAAGGG-------------- .(((.....)))(((((((..((((((((.......))))...))))...((.((((------((((((....))))))--))))))...))))))).-------------- ( -34.40, z-score = -1.99, R) >droPer1.super_2 7102767 112 - 9036312 AGGGCUCUGCAAUUUUUGCGUGUCCGCACGUACACGAGACUCGGGAGUCGGGACUCGGAGCACGGGACUCGGGACUCGCGACUGGACUCGAGAAGGAGGAGGGGCAGGACCA ..((..((((..((((((((((....)))))...((((.((((.((((((((.((((.....)))).)))..))))).)))..)..))))......)))))..))))..)). ( -42.10, z-score = -0.42, R) >consensus AUUGGUGAACAAUUUUUGCGUGUCCGCACGUACACGGUGUUUAGGACUUAGGAUCCG______GGGACUUGGGAGUCCU__CGGACCACGGCAAAGGG______________ ..(((((((((.....((((((....)))))).....)))))((((((((((.(((.......))).)))..))))))).....))))........................ (-24.16 = -22.92 + -1.24)
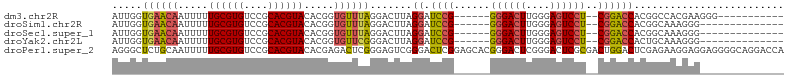

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:53 2011