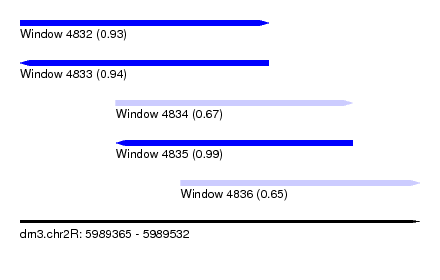
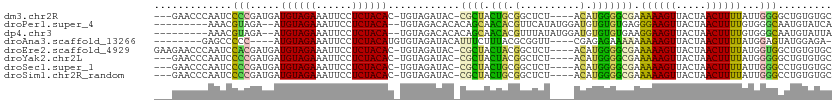
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,989,365 – 5,989,532 |
| Length | 167 |
| Max. P | 0.989713 |

| Location | 5,989,365 – 5,989,469 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.38 |
| Shannon entropy | 0.50599 |
| G+C content | 0.43228 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -8.42 |
| Energy contribution | -9.00 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
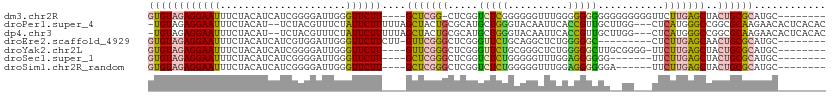
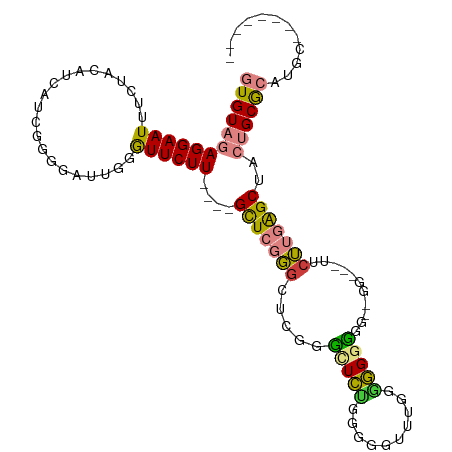
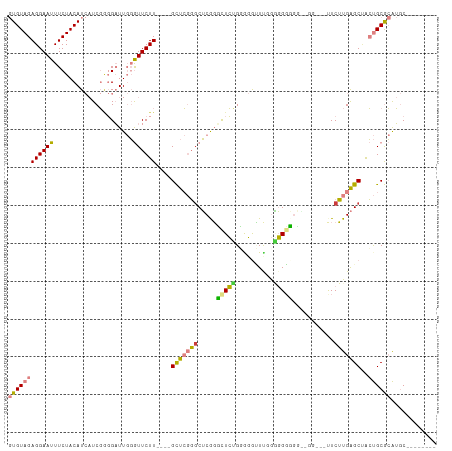
>dm3.chr2R 5989365 104 + 21146708 GCACACAGCCCCAAUAAAAGUUAGUAACUUUUUCGCCCCAUGU----AGAGCCGCAGUAGCG-GUAUCUACA-GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUC--- ((.....))((((((((((((.....))))))...((((.(((----((((((((....)))-)).))))))-((((((((....))))))))....))))))))))...--- ( -39.20, z-score = -5.19, R) >droPer1.super_4 4927170 100 + 7162766 UGAUACAUUGCCCACAAAAGUUAGUAACUUCCCUCACACACAUCCAUAUGAACGUGUUGCUGUGUGUCUACA--UGUAGAGGAAUUUCUACAU--UCUACGUUU--------- .(((((((.((.(((....((.((........)).))...(((....)))...)))..)).)))))))...(--(((((((....))))))))--.........--------- ( -18.70, z-score = -0.32, R) >dp4.chr3 9602207 100 + 19779522 UAAUACAUUGCCCACAAAAGUUAGUAACUUCCUUCACACACAUCCAUAUAAACGUGUUGCUGUGUGUCUACA--UGUAGAGGAAUUUCUACAU--UCUACGUUU--------- .......................(((.........(((((((.(.((((....)))).).)))))))....(--(((((((....))))))))--..)))....--------- ( -16.40, z-score = -0.38, R) >droAna3.scaffold_13266 13847330 96 - 19884421 -UCUCCAUACUCCAUAAAAGUUAGUAACUUUUUUUUUCUCUCG----AACCGCGUAAAGAAAUGUAUCUACACAUGUAGAGGAAUUUCUACAU----GGGGGCUC-------- -........((((((((((((.....))))))...........----..........((((((...(((((....)))))...))))))..))----))))....-------- ( -20.20, z-score = -1.45, R) >droEre2.scaffold_4929 18259364 107 - 26641161 GCACACAGCCACCAUAAAAGUUAGUAACUUUUUCGCCCCAUGU----AGAGCCGUAGUAGCG-GUAUCUACA-GUGUAGAGGAAUUUCUACAUCAUCGUGGAUUGGGUUCUUC ..((((.((......((((((.....))))))..))....(((----((((((((....)))-)).))))))-)))).(((((((((((((......)))))...)))))))) ( -29.30, z-score = -2.23, R) >droYak2.chr2L 18622247 104 + 22324452 GCACACAGCCCCCAUAAAAGUUAGUAACUUUUUCGCCCCAUGU----AGAGCCGUAGUAGCG-GUAUCUACA-GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUC--- ......(((((....((((((.....))))))...((((.(((----((((((((....)))-)).))))))-((((((((....))))))))....))))...))))).--- ( -35.40, z-score = -3.85, R) >droSec1.super_1 3604928 104 + 14215200 GCACACAGGCCCAAUAAAAGUUAGUAACUUUUUCGCCCCAUGU----AGAGCCGCAGUAGCG-GUAUCUACA-GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUC--- .......((((((((((((((.....))))))...((((.(((----((((((((....)))-)).))))))-((((((((....))))))))....)))))))))))).--- ( -41.10, z-score = -5.64, R) >droSim1.chr2R_random 1779547 104 + 2996586 GCACACAGGCCCAAUAAAAGUUAGUAACUUUUUCGCCCCAUGU----AGAGCCGCAGUAGCG-GUAUCUACA-GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUC--- .......((((((((((((((.....))))))...((((.(((----((((((((....)))-)).))))))-((((((((....))))))))....)))))))))))).--- ( -41.10, z-score = -5.64, R) >consensus GCACACAGCCCCCAUAAAAGUUAGUAACUUUUUCGCCCCAUGU____AGAGCCGUAGUAGCG_GUAUCUACA_GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUC___ .........(((...((((((.....)))))).........................................((((((((....))))))))....)))............. ( -8.42 = -9.00 + 0.58)
| Location | 5,989,365 – 5,989,469 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Shannon entropy | 0.50599 |
| G+C content | 0.43228 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -11.73 |
| Energy contribution | -9.82 |
| Covariance contribution | -1.90 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5989365 104 - 21146708 ---GAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACAC-UGUAGAUAC-CGCUACUGCGGCUCU----ACAUGGGGCGAAAAAGUUACUAACUUUUAUUGGGGCUGUGUGC ---...(((((((((((....((((((......)))))).-((((((..(-(((....)))).)))----))))))))...((((((.....))))))))))))......... ( -35.70, z-score = -4.02, R) >droPer1.super_4 4927170 100 - 7162766 ---------AAACGUAGA--AUGUAGAAAUUCCUCUACA--UGUAGACACACAGCAACACGUUCAUAUGGAUGUGUGUGAGGGAAGUUACUAACUUUUGUGGGCAAUGUAUCA ---------..((((...--(((((((......))))))--)((...(((...(((.(((((((....))))))))))...((((((.....))))))))).)).)))).... ( -24.50, z-score = -0.69, R) >dp4.chr3 9602207 100 - 19779522 ---------AAACGUAGA--AUGUAGAAAUUCCUCUACA--UGUAGACACACAGCAACACGUUUAUAUGGAUGUGUGUGAAGGAAGUUACUAACUUUUGUGGGCAAUGUAUUA ---------..((((...--(((((((......))))))--)((...(((...(((.(((((((....))))))))))...((((((.....))))))))).)).)))).... ( -22.10, z-score = -0.45, R) >droAna3.scaffold_13266 13847330 96 + 19884421 --------GAGCCCCC----AUGUAGAAAUUCCUCUACAUGUGUAGAUACAUUUCUUUACGCGGUU----CGAGAGAAAAAAAAAGUUACUAACUUUUAUGGAGUAUGGAGA- --------(((((...----(((((((......)))))))(((((((........)))))))))))----)..................(((((((.....)))).)))...- ( -21.90, z-score = -1.66, R) >droEre2.scaffold_4929 18259364 107 + 26641161 GAAGAACCCAAUCCACGAUGAUGUAGAAAUUCCUCUACAC-UGUAGAUAC-CGCUACUACGGCUCU----ACAUGGGGCGAAAAAGUUACUAACUUUUAUGGUGGCUGUGUGC ......((((...........((((((......)))))).-((((((..(-((......))).)))----)))))))(((....((((((((.......))))))))...))) ( -29.10, z-score = -2.02, R) >droYak2.chr2L 18622247 104 - 22324452 ---GAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACAC-UGUAGAUAC-CGCUACUACGGCUCU----ACAUGGGGCGAAAAAGUUACUAACUUUUAUGGGGGCUGUGUGC ---(..((((..(((((....((((((......)))))).-((((((..(-((......))).)))----))))))))...((((((.....)))))).))))..)....... ( -31.70, z-score = -2.51, R) >droSec1.super_1 3604928 104 - 14215200 ---GAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACAC-UGUAGAUAC-CGCUACUGCGGCUCU----ACAUGGGGCGAAAAAGUUACUAACUUUUAUUGGGCCUGUGUGC ---...(((((((((((....((((((......)))))).-((((((..(-(((....)))).)))----))))))))...((((((.....))))))))))))......... ( -34.80, z-score = -4.17, R) >droSim1.chr2R_random 1779547 104 - 2996586 ---GAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACAC-UGUAGAUAC-CGCUACUGCGGCUCU----ACAUGGGGCGAAAAAGUUACUAACUUUUAUUGGGCCUGUGUGC ---...(((((((((((....((((((......)))))).-((((((..(-(((....)))).)))----))))))))...((((((.....))))))))))))......... ( -34.80, z-score = -4.17, R) >consensus ___GAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACAC_UGUAGAUAC_CGCUACUACGGCUCU____ACAUGGGGCGAAAAAGUUACUAACUUUUAUGGGGGCUGUGUGC .............(((.....((((((......))))))............((((.(((.(..........).))))))).((((((.....))))))...)))......... (-11.73 = -9.82 + -1.90)
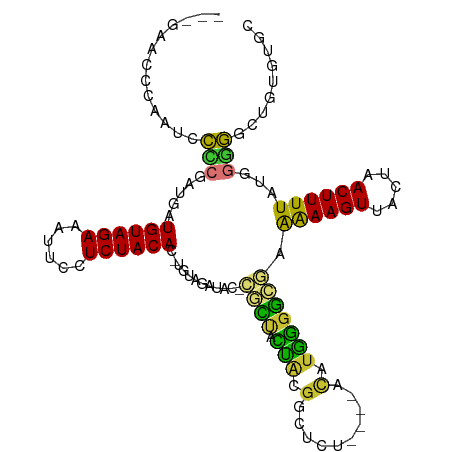
| Location | 5,989,405 – 5,989,504 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.28 |
| Shannon entropy | 0.52376 |
| G+C content | 0.50670 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -12.06 |
| Energy contribution | -10.61 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5989405 99 + 21146708 --------UGUAGAGCCGCAGUAGCG-GUAUCUACAGUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU---GCUCGG-CUCGGUCUCGGGGGGUUUGGGGGGGGG --------(((((((((((....)))-)).))))))...........(..(((.(((..(.(((((((((((...---....))-))))))))).)..))).)))..).... ( -35.20, z-score = -2.25, R) >droPer1.super_4 4927206 103 + 7162766 CACACAUCCAUAUGAACGUGUUGCUGUGUGUCUACA-UGUAGAGGAAUUUCUACAU--UCUACGUUUCUAUUCUU----UUUAGCUACUGCGCAUGCUGGGUACAAUUCA-- ((((((.(.((((....)))).).))))))((((((-(((((((((((....((..--.....))....))))))----))..((....))))))).)))).........-- ( -21.50, z-score = -0.31, R) >dp4.chr3 9602243 103 + 19779522 CACACAUCCAUAUAAACGUGUUGCUGUGUGUCUACA-UGUAGAGGAAUUUCUACAU--UCUACGUUUCUAUUCUU----UUUAGCUACUGCGCAUGCUGGGUACAAUUCA-- ((((((.(.((((....)))).).))))))((((((-(((((((((((....((..--.....))....))))))----))..((....))))))).)))).........-- ( -20.60, z-score = -0.43, R) >droEre2.scaffold_4929 18259404 101 - 26641161 --------UGUAGAGCCGUAGUAGCG-GUAUCUACAGUGUAGAGGAAUUUCUACAUCAUCGUGGAUUGGGUUCUUCUUGUUCGGGCUCGGGUUCUGCAGGCUCUGGGGGC-- --------(.(((((((((((.(((.-...(((((.((((((((....))))))))....)))))((((((((.........))))))))))))))).))))))).)...-- ( -34.20, z-score = -1.75, R) >droYak2.chr2L 18622287 100 + 22324452 --------UGUAGAGCCGUAGUAGCG-GUAUCUACAGUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU---GUUCGGGCUCGGGUUCUGCGGGCUCUGGGGGCUU --------(((((((((((....)))-)).))))))((((((((....))))))))...((.(((((((((((..---....))))))))..))).))((((.....)))). ( -34.40, z-score = -1.56, R) >droSec1.super_1 3604968 100 + 14215200 --------UGUAGAGCCGCAGUAGCG-GUAUCUACAGUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU---GCUCGGGCUCGGUCUCUGGGGGUUUGGAGGGGGG --------(((((((((((....)))-)).))))))...........((((((.(((..((((((((((((((..---....))))))))))))))..))).)))))).... ( -40.60, z-score = -3.75, R) >droSim1.chr2R_random 1779587 100 + 2996586 --------UGUAGAGCCGCAGUAGCG-GUAUCUACAGUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU---GCUCGGGCUCGGUCUCUGGGGGUUUGGAGGGGGG --------(((((((((((....)))-)).))))))...........((((((.(((..((((((((((((((..---....))))))))))))))..))).)))))).... ( -40.60, z-score = -3.75, R) >consensus ________UGUAGAGCCGUAGUAGCG_GUAUCUACAGUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU___GUUCGGGCUCGGUCUCUGCGGGUUUGGAGGGG__ ...............((.(((.(((...........((((((((....)))))))).........(((((..(.........)..))))).........)))))).)).... (-12.06 = -10.61 + -1.44)

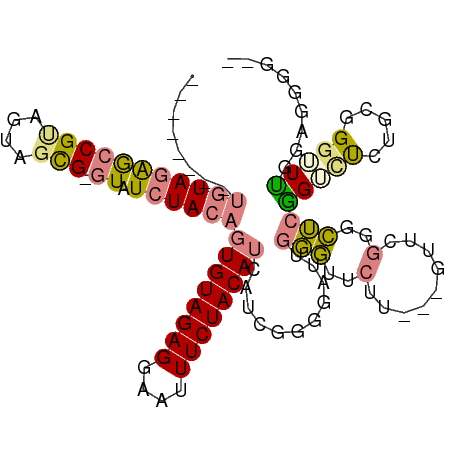
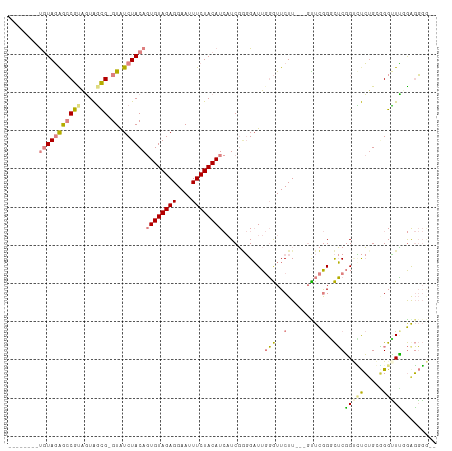
| Location | 5,989,405 – 5,989,504 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.28 |
| Shannon entropy | 0.52376 |
| G+C content | 0.50670 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.44 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5989405 99 - 21146708 CCCCCCCCCAAACCCCCCGAGACCGAG-CCGAGC---AAGAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACACUGUAGAUAC-CGCUACUGCGGCUCUACA-------- ........................(((-(((...---....................((((((......))))))..((((....-..))))..))))))....-------- ( -20.00, z-score = -3.59, R) >droPer1.super_4 4927206 103 - 7162766 --UGAAUUGUACCCAGCAUGCGCAGUAGCUAAA----AAGAAUAGAAACGUAGA--AUGUAGAAAUUCCUCUACA-UGUAGACACACAGCAACACGUUCAUAUGGAUGUGUG --................(((...((..(((..----.....)))..))((...--(((((((......))))))-)....)).....)))((((((((....)))))))). ( -22.00, z-score = -0.95, R) >dp4.chr3 9602243 103 - 19779522 --UGAAUUGUACCCAGCAUGCGCAGUAGCUAAA----AAGAAUAGAAACGUAGA--AUGUAGAAAUUCCUCUACA-UGUAGACACACAGCAACACGUUUAUAUGGAUGUGUG --................(((...((..(((..----.....)))..))((...--(((((((......))))))-)....)).....)))((((((((....)))))))). ( -19.70, z-score = -0.30, R) >droEre2.scaffold_4929 18259404 101 + 26641161 --GCCCCCAGAGCCUGCAGAACCCGAGCCCGAACAAGAAGAACCCAAUCCACGAUGAUGUAGAAAUUCCUCUACACUGUAGAUAC-CGCUACUACGGCUCUACA-------- --......(((((((((((....((....))..........................((((((......))))))))))))....-((......)))))))...-------- ( -19.80, z-score = -2.31, R) >droYak2.chr2L 18622287 100 - 22324452 AAGCCCCCAGAGCCCGCAGAACCCGAGCCCGAAC---AAGAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACACUGUAGAUAC-CGCUACUACGGCUCUACA-------- ........((((((....................---....................((((((......))))))..((((....-..))))...))))))...-------- ( -18.50, z-score = -2.26, R) >droSec1.super_1 3604968 100 - 14215200 CCCCCCUCCAAACCCCCAGAGACCGAGCCCGAGC---AAGAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACACUGUAGAUAC-CGCUACUGCGGCUCUACA-------- .....(((..........)))...(((((...((---(...................((((((......))))))..((((....-..))))))))))))....-------- ( -19.40, z-score = -2.77, R) >droSim1.chr2R_random 1779587 100 - 2996586 CCCCCCUCCAAACCCCCAGAGACCGAGCCCGAGC---AAGAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACACUGUAGAUAC-CGCUACUGCGGCUCUACA-------- .....(((..........)))...(((((...((---(...................((((((......))))))..((((....-..))))))))))))....-------- ( -19.40, z-score = -2.77, R) >consensus __CCCCUCCAAACCCGCAGAGACCGAGCCCGAAC___AAGAACCCAAUCCCCGAUGAUGUAGAAAUUCCUCUACACUGUAGAUAC_CGCUACUACGGCUCUACA________ ........................(((((............................((((((......))))))..((((..........)))))))))............ (-11.13 = -11.44 + 0.31)
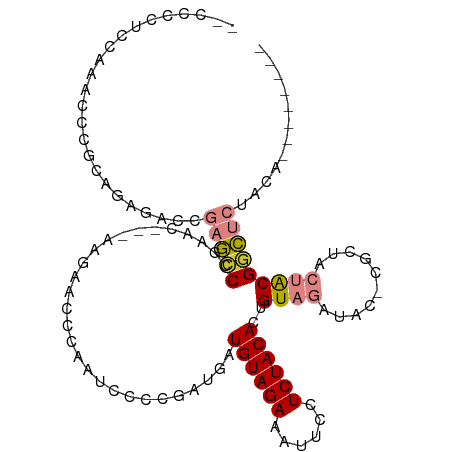
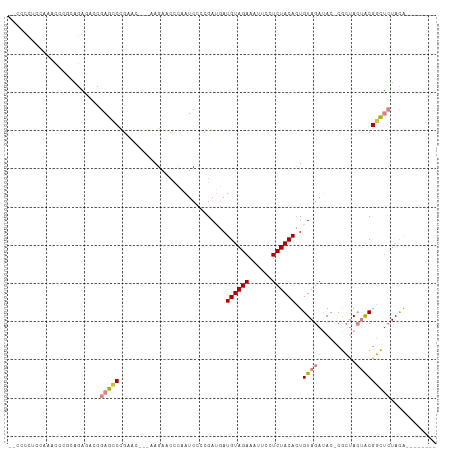
| Location | 5,989,432 – 5,989,532 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.58946 |
| G+C content | 0.54195 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -12.71 |
| Energy contribution | -11.53 |
| Covariance contribution | -1.18 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5989432 100 + 21146708 GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU----GCUCGG-CUCGGUCUCGGGGGGUUUGGGGGGGGGGGGGGGGUUCUUGAGCUACUGCGCAUGC-------- ((((((.....(..(((.(((..(.(((((((((((...----....))-))))))))).)..))).)))..).........((((....)))).))))))....-------- ( -30.10, z-score = -0.01, R) >droPer1.super_4 4927242 107 + 7162766 -UGUAGAGGAAUUUCUACAU--UCUACGUUUCUAUUCUUUUUAGCUACUGCGCAUGCUGGGUACAAUUCACCGUUGCUUGG---CUCAUGGGCCGGCGCAAGAACACUCACAC -(((((((....)))))))(--(((..((..(((.......)))..))(((((..((..(((.......)))...)).(((---((....))))))))))))))......... ( -26.50, z-score = -0.98, R) >dp4.chr3 9602279 107 + 19779522 -UGUAGAGGAAUUUCUACAU--UCUACGUUUCUAUUCUUUUUAGCUACUGCGCAUGCUGGGUACAAUUCACCGUUGCUUGG---CUCAUGGGCCGGCGCAAGAACACUCACAC -(((((((....)))))))(--(((..((..(((.......)))..))(((((..((..(((.......)))...)).(((---((....))))))))))))))......... ( -26.50, z-score = -0.98, R) >droEre2.scaffold_4929 18259431 95 - 26641161 GUGUAGAGGAAUUUCUACAUCAUCGUGGAUUGGGUUCUUCUU-GUUCGGGCUCGGGUUCUGCAGGCUCUGGGGGC---------CUCUUGAGCAACUGCGCAUGC-------- ((((((((((((((((((......)))))...))))))))((-(((((((...(((((((.(((...))))))))---------))))))))))).)))))....-------- ( -33.30, z-score = -1.42, R) >droYak2.chr2L 18622314 100 + 22324452 GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU----GUUCGGGCUCGGGUUCUGCGGGCUCUGGGGGCUUGCGGGG-UUCUUGAGCUACUGCGCAUGC-------- (((((((((((((((..(......)..))...)))))))----.....((((((((.((((((((((.....)))))))))).-..)))))))).))))))....-------- ( -38.50, z-score = -2.44, R) >droSec1.super_1 3604995 94 + 14215200 GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU----GCUCGGGCUCGGUCUCUGGGGGUUUGGAGGGGGG-------UUCUUGAGCUACUGCGCAUGC-------- ((((((((((((..((.((...(((((((((((((((..----....))))))))))))))).....))...))..)-------)))))......))))))....-------- ( -35.70, z-score = -2.65, R) >droSim1.chr2R_random 1779614 95 + 2996586 GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU----GCUCGGGCUCGGUCUCUGGGGGUUUGGAGGGGGGA------UUCUUGAGCUACUGCGCAUGC-------- ((((((((((((..((.((...(((((((((((((((..----....))))))))))))))).....))....))..)------)))))......))))))....-------- ( -34.80, z-score = -2.28, R) >consensus GUGUAGAGGAAUUUCUACAUCAUCGGGGAUUGGGUUCUU____GCUCGGGCUCGGGCUCUGGGGGUUUGGGGGGGGG__GG___UUCUUGAGCUACUGCGCAUGC________ ((((((((....)))))))).......................((....(((((((((((..........)))))............))))))....)).............. (-12.71 = -11.53 + -1.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:48 2011