| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,950,380 – 5,950,476 |
| Length | 96 |
| Max. P | 0.822691 |

| Location | 5,950,380 – 5,950,476 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.64372 |
| G+C content | 0.45694 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -12.35 |
| Energy contribution | -11.97 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
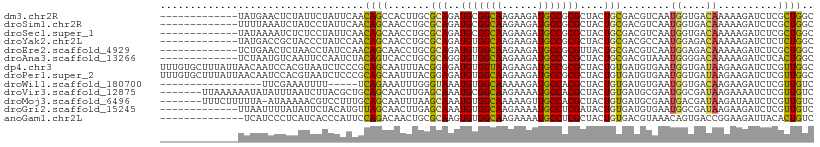
>dm3.chr2R 5950380 96 + 21146708 -------------UAUGAACUCUAUUCUAUUCAACAGCCACUUGCGCAGAUGCGGCAAGAAGAUGCCGCGCUACUGCGACGUCAAUGGUGACAAAAAGAUCUCGCUGGC -------------.....................((((......(((((.(((((((......)))))))...)))))..((((....))))...........)))).. ( -28.10, z-score = -1.25, R) >droSim1.chr2R 4580399 96 + 19596830 -------------UUUUAAAUCUAUCCUAUUCAACAGCAACCUGCGCAGAUGCGGCAAGAAGAUGCCGCGCUACUGCGACGUCAAUGGUGACAAAAAGAUCUCGCUGGC -------------.....................((((......(((((.(((((((......)))))))...)))))..((((....))))...........)))).. ( -29.00, z-score = -1.83, R) >droSec1.super_1 3562797 96 + 14215200 -------------UAUAAAAUCUCUCCUAUUCAACAGCAACCUGCGCAGAUGCGGCAAGAAGAUGCCGCGCUACUGCGACGUCAAUGGUGACAAAAAGAUCUCGCUGGC -------------.....................((((......(((((.(((((((......)))))))...)))))..((((....))))...........)))).. ( -29.00, z-score = -1.95, R) >droYak2.chr2L 18583065 96 + 22324452 -------------UAUGACCGCUACCCUAUCCAACAGCAACCUGCGCAGAUGUGGCAAGAAGAUGCCGCGCUACUGCGACGCCAAUGGAGACAAAAAGAUCUCUCUGGC -------------.......(((............)))......(((((.(((((((......)))))))...)))))..((((..(((((........))))).)))) ( -26.90, z-score = -1.50, R) >droEre2.scaffold_4929 18220157 96 - 26641161 -------------UCUGAACUCUAACCUAUCCAACAGCAACCUGCGCAGAUGUGGCAAGAAGAUGCCGCGUUACUGCGACGUCAAUGGAGACAAAAAGAUCUCGCUGGC -------------.....................((((......(((((((((((((......))))))))..))))).........((((........)))))))).. ( -28.30, z-score = -1.85, R) >droAna3.scaffold_13266 13805894 96 - 19884421 -------------UCUAAUGUCAAUUCCAAUCUACAGUCACCUGCGCAGGUGUGGCAAGAAGAUGCCCCGCUACUGCGACGUAAAUGGGGACAAAAAGAUCUCACUGGC -------------(((..((((...((((.....(((....)))((((((((.((((......)))).)))..))))).......))))))))...))).......... ( -22.90, z-score = 0.16, R) >dp4.chr3 6163752 109 + 19779522 UUUGUGCUUUAUUAACAAUCCACGUAAUCUCCCGCAGCAAUUUACGGAGAUGUGGUAAGAAGAUGCCGCGCUACUGUGAUGUGAAUGGUGAUAAGAAGAUCUCGUUGGC .((((.........)))).(((((..(((((.(((..((..((((((((.(((((((......))))))))).))))))..))....)))....).))))..)).))). ( -24.80, z-score = 0.20, R) >droPer1.super_2 6377089 109 + 9036312 UUUGUGCUUUAUUAACAAUCCACGUAAUCUCCCGCAGCAAUUUACGGAGAUGUGGCAAGAAGAUGCCGCGCUACUGUGAUGUGAAUGGUGAUAAGAAGAUCUCGUUGGC .((((.........)))).(((((..(((((.(((..((..((((((((.(((((((......))))))))).))))))..))....)))....).))))..)).))). ( -26.80, z-score = -0.16, R) >droWil1.scaffold_180700 5396318 87 + 6630534 -----------------UUCGAAAUUUU-----UCAGAAAUUUGGGUAAAUGUGGCAAAAAGAUGCCACGCUACUGUGAUGUGAAUGGUGACAAGAAGAUCUCGUUGUC -----------------..(((...(((-----((.........((((..(((((((......))))))).))))..........((....)).)))))..)))..... ( -19.30, z-score = -0.87, R) >droVir3.scaffold_12875 171695 102 - 20611582 -------UUAAAAAAUAUAUUUAAUCUUACGCUGCAGCAACUUGAGCAAAUGCGGCAAGAAAAUGCCACGCUACUGUGAUGCGAAUGGCGAUAAGAAAAUCUCGUUGUC -------.................((((((((((..(((......((....))((((......)))).(((....))).)))...))))).)))))............. ( -21.60, z-score = -0.35, R) >droMoj3.scaffold_6496 5698036 101 + 26866924 -------UUUCUUUUUA-AUAAAAACGUCCUUUGCAGCAAUUUAAGCAAAUGUGGCAAAAAGUUGCCACGCUACUGUGAUGCGAAUGACGAUAAGAUAAUCUCGUUGUC -------..........-.......((((.((((((.((.....(((....(((((((....))))))))))....)).)))))).))))....((((((...)))))) ( -26.10, z-score = -2.57, R) >droGri2.scaffold_15245 11109287 96 + 18325388 -------------UUAAUUUUAUAUUCUACAUGUUAGCAACUUGAGCAAAUGUGGCAAGAAAAUGCCUCGAUACUGUGAUGUGAAUGGCGAUAAGAAGAUCUCGUUGUC -------------.....((((((((..(((((((.((.......))....(.((((......)))).))))).))))))))))).(((((..((.....))..))))) ( -17.70, z-score = 0.19, R) >anoGam1.chr2L 2492854 95 - 48795086 --------------UCAUCCCUCAUCACCCAUUCCAGACAACUGCGCAAGUGUGGCAAGAAAAUGCCUCGCUACUGUGACGUAAACAGUGACCGGAAGAUUACACUGUC --------------......................((((((..((((((((.((((......)))).))))..))))..)).....((((.(....).))))..)))) ( -22.00, z-score = -0.89, R) >consensus _____________UAUAAACUCUAUCCUAUUCAACAGCAACUUGCGCAGAUGUGGCAAGAAGAUGCCGCGCUACUGUGACGUGAAUGGUGACAAGAAGAUCUCGCUGGC ..................................((((.............((((((......))))))((....))..........................)))).. (-12.35 = -11.97 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:38 2011