| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,923,137 – 5,923,244 |
| Length | 107 |
| Max. P | 0.947596 |

| Location | 5,923,137 – 5,923,239 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Shannon entropy | 0.27030 |
| G+C content | 0.46447 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
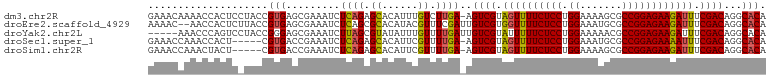
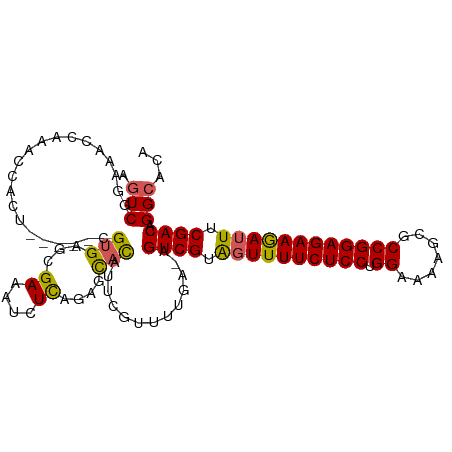
>dm3.chr2R 5923137 102 - 21146708 GAAACAAAACCACUCCUACCGUGAGCGAAAUCUCAGAGCACAUUUGUCUUGA-AGUCGUAGUUUUCUCCUGGAAAAGCGCCGGAGAAGAUUUCGACAGGCACA ..........(((.......))).((......((((..((....))..))))-.((((.((((((((((.((.......)))))))))))).))))..))... ( -27.10, z-score = -1.18, R) >droEre2.scaffold_4929 18193589 101 + 26641161 AAAAC--AACCACUCUUACCGUGAGCGAAAUCUCAGCGCACAUACGUUUCGAUUGUCGUGGUUUUCUCCUGGAAAUGCGCCGGAGAAGAUUUCGACAGGCACA .....--...........(((((.((((....)).)).)))............(((((..(((((((((.((.......)))))))))))..))))))).... ( -28.40, z-score = -1.20, R) >droYak2.chr2L 18554877 98 - 22324452 -----AAACCCAGUCCUACCGGGAGCGAAAUCUUAGCGUAUAUUUGUUUUGAUUGUCGUAUUUUUCUCCUGGAAAAACGCCGGAGAAGAUUUCGACAGGCACA -----...(((.(.....).))).((..((((..((((......))))..))))((((...((((((((.((.......))))))))))...))))..))... ( -29.00, z-score = -2.21, R) >droSec1.super_1 3537117 97 - 14215200 GAAACCAAACCACU-----CGUGACCGAAAUCUCAGAGCACAUUCGUUUUGA-AGUCGUAGUUUUCUCCUGGAAAUGCGCCGGAGAAAAUUUCGACAGGCACA ..............-----.(((.((......(((((((......)))))))-.((((.((((((((((.((.......)))))))))))).)))).))))). ( -32.50, z-score = -3.59, R) >droSim1.chr2R 4553308 97 - 19596830 GAAACCAAACUACU-----CGUGACCGAAAUCUCAGAGCACAUUCGUUUUGA-AGUCGUAGUUUUCUCCUGGAAAAGCGCCGGAGAAGAUUUCGACAGGCACA ..............-----.(((.((......(((((((......)))))))-.((((.((((((((((.((.......)))))))))))).)))).))))). ( -32.60, z-score = -3.36, R) >consensus GAAACAAAACCACUC_UACCGUGAGCGAAAUCUCAGAGCACAUUCGUUUUGA_AGUCGUAGUUUUCUCCUGGAAAAGCGCCGGAGAAGAUUUCGACAGGCACA ....................(((..((((((..............))))))...((((.((((((((((.((.......)))))))))))).))))...))). (-22.10 = -22.22 + 0.12)
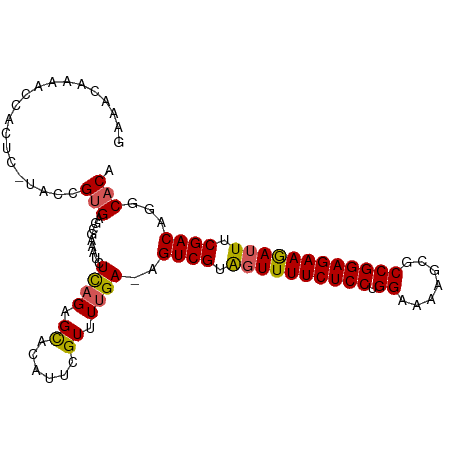
| Location | 5,923,137 – 5,923,244 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Shannon entropy | 0.25797 |
| G+C content | 0.47100 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.14 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.669857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5923137 107 - 21146708 AGUCGGAAACAAAACCACUCCUACCGUGAGCGAAAUCUCAGAGCACAUUUGUCUUGA-AGUCGUAGUUUUCUCCUGGAAAAGCGCCGGAGAAGAUUUCGACAGGCACA .((((....).....(.((((....).))).).....((((..((....))..))))-.((((.((((((((((.((.......)))))))))))).)))).)))... ( -29.30, z-score = -0.89, R) >droEre2.scaffold_4929 18193589 106 + 26641161 AGUCGAAAAC--AACCACUCUUACCGUGAGCGAAAUCUCAGCGCACAUACGUUUCGAUUGUCGUGGUUUUCUCCUGGAAAUGCGCCGGAGAAGAUUUCGACAGGCACA ((((((((..--...(((.......))).(((.........))).......))))))))((((..(((((((((.((.......)))))))))))..))))....... ( -31.20, z-score = -1.24, R) >droYak2.chr2L 18554877 103 - 22324452 ACUCGAAACCCAGUCCUAC-----CGGGAGCGAAAUCUUAGCGUAUAUUUGUUUUGAUUGUCGUAUUUUUCUCCUGGAAAAACGCCGGAGAAGAUUUCGACAGGCACA ..(((...(((.(.....)-----.)))..)))((((..((((......))))..))))((((...((((((((.((.......))))))))))...))))....... ( -29.00, z-score = -1.63, R) >droSec1.super_1 3537117 102 - 14215200 AGUCGGAAACCAAACCACU-----CGUGACCGAAAUCUCAGAGCACAUUCGUUUUGA-AGUCGUAGUUUUCUCCUGGAAAUGCGCCGGAGAAAAUUUCGACAGGCACA (((.((........)))))-----.(((.((......(((((((......)))))))-.((((.((((((((((.((.......)))))))))))).)))).))))). ( -33.50, z-score = -2.84, R) >droSim1.chr2R 4553308 102 - 19596830 AGUCGGAAACCAAACUACU-----CGUGACCGAAAUCUCAGAGCACAUUCGUUUUGA-AGUCGUAGUUUUCUCCUGGAAAAGCGCCGGAGAAGAUUUCGACAGGCACA ....(....).........-----.(((.((......(((((((......)))))))-.((((.((((((((((.((.......)))))))))))).)))).))))). ( -33.50, z-score = -2.58, R) >consensus AGUCGGAAACCAAACCACU_____CGUGAGCGAAAUCUCAGAGCACAUUCGUUUUGA_AGUCGUAGUUUUCUCCUGGAAAAGCGCCGGAGAAGAUUUCGACAGGCACA .(((.....................(((...((....))....))).............((((.((((((((((.((.......)))))))))))).)))).)))... (-22.34 = -22.14 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:36 2011