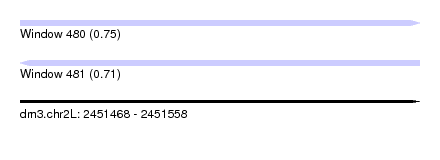
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,451,468 – 2,451,558 |
| Length | 90 |
| Max. P | 0.748575 |

| Location | 2,451,468 – 2,451,558 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.59119 |
| G+C content | 0.45577 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -10.16 |
| Energy contribution | -10.34 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2451468 90 + 23011544 ---------AGUG-UGUGUCUUCAGCUGGGUGUGUGGAAUGUCAACUGACGGGUUGUAAAGGGAAACCCUGAAAUCCGAACGGCC---AGCCAAAGCAAAUAA ---------..((-(.((.(((..(((((.(((.((((..(((....)))(((((.........))))).....)))).))).))---)))..))))).))). ( -30.00, z-score = -2.22, R) >droSim1.chr2L 2411754 90 + 22036055 ---------AGUG-UGUGUCUUCAGCUGGGUGUGUGGAAUGUCAACUGACGGGUUGUAAAGGGAAACCCUGAAAUCCGAACGGCC---AGCCAAAGCAAAUAA ---------..((-(.((.(((..(((((.(((.((((..(((....)))(((((.........))))).....)))).))).))---)))..))))).))). ( -30.00, z-score = -2.22, R) >droSec1.super_5 625003 74 + 5866729 --------------------------AGUGUGUGUGGAAUGUCAACAGACGGGUUGUAAAGGGAAACCCUGAAAUCCGAACGGCC---AGCCAAAGCAAAUAA --------------------------..(((...(((...(((......((((((....((((...))))..))))))...))).---..)))..)))..... ( -18.50, z-score = -0.77, R) >droYak2.chr2L 2440812 90 + 22324452 ---------AGUG-UGUGUCUUCAGCUGGGUGUGUGGAAUGUCAACUGACGGGUUGUAAAGGGAAACCCUGAAAUCCGAACGGCC---AGCCAAAGCAAAUAA ---------..((-(.((.(((..(((((.(((.((((..(((....)))(((((.........))))).....)))).))).))---)))..))))).))). ( -30.00, z-score = -2.22, R) >droEre2.scaffold_4929 2495721 90 + 26641161 ---------AGUG-UGUGUCUUCAGCUGGGUGUGUGGAAUGUCAACUGACGGGUUGUAAAGGGAAACCCUGAAAUCCGAACAGCC---AGCCAAAGCAAAUAA ---------..((-(.((.(((..(((((.(((.((((..(((....)))(((((.........))))).....)))).))).))---)))..))))).))). ( -30.70, z-score = -2.65, R) >droAna3.scaffold_12913 191567 100 - 441482 ---AGUGCUGGUGCUGAAGCCGAUGCUGGGUGUUUGGAAUGUCAACUGACGGGUUGUAAAGGGAAACCCUGAAAUUCGAAUAGCCGCCAGCUAAAGCAAAUAA ---..((((...((((.(((....)))((.((((((((..(((....)))(((((.........))))).....)))))))).))..))))...))))..... ( -32.30, z-score = -2.12, R) >droWil1.scaffold_180703 2977729 96 + 3946847 AGGUGUGCGUGUG-UGUGUGUGCGACUUUUGGAAUGGAAUGUCAACUGACAAGUUGUAAAGGGAAACCCUGAAAUUCGAACAGCC------AAAAAUAUAUAA ..((((...((.(-(.(((...(((.(((..(.(..(..((((....))))..)..)...((....)))..))).))).))))))------)...)))).... ( -20.90, z-score = -0.52, R) >droGri2.scaffold_15252 5192514 73 - 17193109 -------UGUGUG-UGCGAGUGUGUGUGUCUUGCUGGAAUGUCAACUAACAAGUUGAAAUAAAAAAUUGUUAAACAUCAAU---------------------- -------((.(((-((((((.........))))).......((((((....))))))................))))))..---------------------- ( -11.30, z-score = 0.06, R) >consensus _________AGUG_UGUGUCUUCAGCUGGGUGUGUGGAAUGUCAACUGACGGGUUGUAAAGGGAAACCCUGAAAUCCGAACGGCC___AGCCAAAGCAAAUAA ............................(((....((...(((....)))(((((.....((....))....)))))......))....)))........... (-10.16 = -10.34 + 0.17)

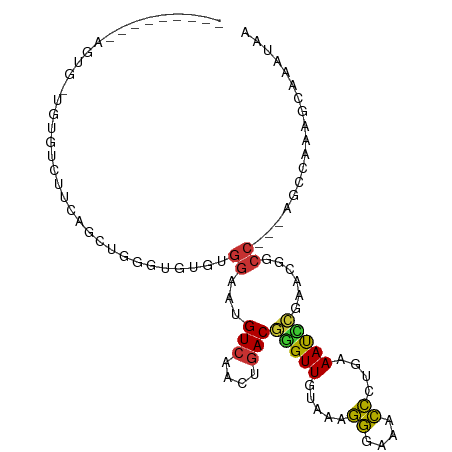
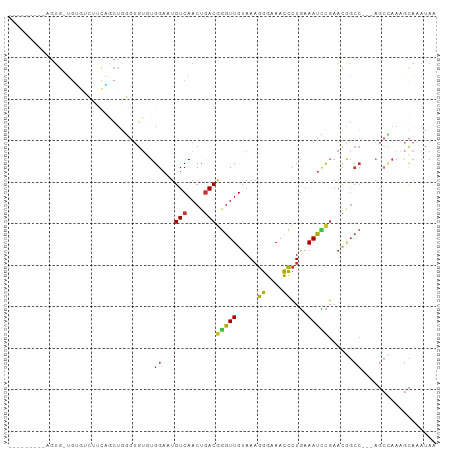
| Location | 2,451,468 – 2,451,558 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.59119 |
| G+C content | 0.45577 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -7.13 |
| Energy contribution | -7.17 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 2451468 90 - 23011544 UUAUUUGCUUUGGCU---GGCCGUUCGGAUUUCAGGGUUUCCCUUUACAACCCGUCAGUUGACAUUCCACACACCCAGCUGAAGACACA-CACU--------- .....(((((..(((---((..((..(((.....(((((.........)))))(((....)))..)))..))..)))))..))).))..-....--------- ( -27.10, z-score = -3.24, R) >droSim1.chr2L 2411754 90 - 22036055 UUAUUUGCUUUGGCU---GGCCGUUCGGAUUUCAGGGUUUCCCUUUACAACCCGUCAGUUGACAUUCCACACACCCAGCUGAAGACACA-CACU--------- .....(((((..(((---((..((..(((.....(((((.........)))))(((....)))..)))..))..)))))..))).))..-....--------- ( -27.10, z-score = -3.24, R) >droSec1.super_5 625003 74 - 5866729 UUAUUUGCUUUGGCU---GGCCGUUCGGAUUUCAGGGUUUCCCUUUACAACCCGUCUGUUGACAUUCCACACACACU-------------------------- ...........((..---..))....(((.....(((((.........)))))(((....)))..))).........-------------------------- ( -13.50, z-score = 0.16, R) >droYak2.chr2L 2440812 90 - 22324452 UUAUUUGCUUUGGCU---GGCCGUUCGGAUUUCAGGGUUUCCCUUUACAACCCGUCAGUUGACAUUCCACACACCCAGCUGAAGACACA-CACU--------- .....(((((..(((---((..((..(((.....(((((.........)))))(((....)))..)))..))..)))))..))).))..-....--------- ( -27.10, z-score = -3.24, R) >droEre2.scaffold_4929 2495721 90 - 26641161 UUAUUUGCUUUGGCU---GGCUGUUCGGAUUUCAGGGUUUCCCUUUACAACCCGUCAGUUGACAUUCCACACACCCAGCUGAAGACACA-CACU--------- .....(((((..(((---((.(((..(((.....(((((.........)))))(((....)))..)))..))).)))))..))).))..-....--------- ( -29.60, z-score = -3.91, R) >droAna3.scaffold_12913 191567 100 + 441482 UUAUUUGCUUUAGCUGGCGGCUAUUCGAAUUUCAGGGUUUCCCUUUACAACCCGUCAGUUGACAUUCCAAACACCCAGCAUCGGCUUCAGCACCAGCACU--- .....((((..((((((..(((............(((((.........)))))(((....))).............))).))))))..))))........--- ( -23.50, z-score = -1.63, R) >droWil1.scaffold_180703 2977729 96 - 3946847 UUAUAUAUUUUU------GGCUGUUCGAAUUUCAGGGUUUCCCUUUACAACUUGUCAGUUGACAUUCCAUUCCAAAAGUCGCACACACA-CACACGCACACCU ......((((((------((..((..((((...((((...))))...(((((....)))))..)))).)).)))))))).((.......-.....))...... ( -12.70, z-score = -0.07, R) >droGri2.scaffold_15252 5192514 73 + 17193109 ----------------------AUUGAUGUUUAACAAUUUUUUAUUUCAACUUGUUAGUUGACAUUCCAGCAAGACACACACACUCGCA-CACACA------- ----------------------..((.((((((((((.((........)).))))))((((......))))..))))))..........-......------- ( -6.40, z-score = 0.17, R) >consensus UUAUUUGCUUUGGCU___GGCCGUUCGGAUUUCAGGGUUUCCCUUUACAACCCGUCAGUUGACAUUCCACACACCCAGCUGAAGACACA_CACU_________ ..........((((.....))))...........(((((.........)))))(((....)))........................................ ( -7.13 = -7.17 + 0.05)
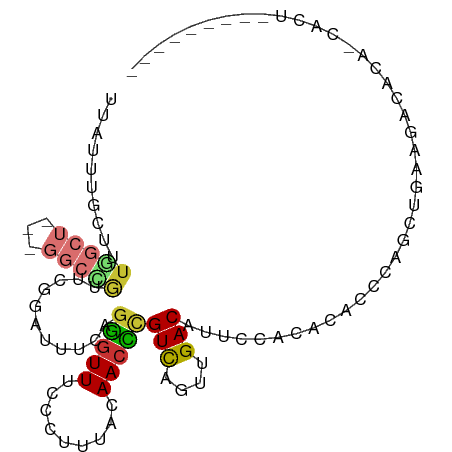
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:19 2011