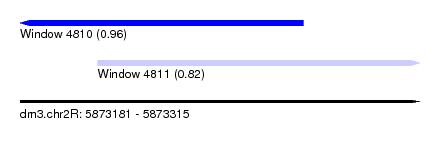
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,873,181 – 5,873,315 |
| Length | 134 |
| Max. P | 0.962911 |

| Location | 5,873,181 – 5,873,276 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.93 |
| Shannon entropy | 0.14516 |
| G+C content | 0.59069 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.16 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
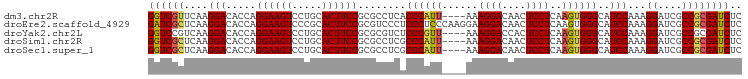
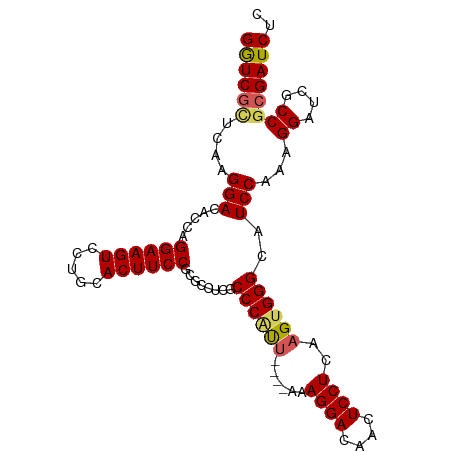
>dm3.chr2R 5873181 95 - 21146708 GGUCGUUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCACCCAUU----AAAGGACAACUCCUCAAGUGGGCAUCCAAAGGAUCGCCGCGAUCUC ((((((....(((...((((....))))....)))(((.....((((((----..((((....))))..)))))).((((...))))))).)))))).. ( -30.40, z-score = -2.10, R) >droEre2.scaffold_4929 18145261 99 + 26641161 GAUCGCUCAAGGACACCAGGAAGUCCCGCACUUCCGCGUCCCUCCCUCCCAAGGAAGGACAACUCCUCAAGUGGGCAUCCAAAGGAUCGCCGCGAUCUC ((((((...((((.....((((((.....))))))..((((.(((.......))).))))...))))......(((((((...)))).))))))))).. ( -37.70, z-score = -3.36, R) >droYak2.chr2L 18501172 95 - 22324452 GGUCCGUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCGUCUCCCGUU----AAAGGACCACUCCUCAAGUGGGCAUCCAAAGGAUCGCCGCGAUCUC .((((.....))))....((((((.....)))))).((((....((...----...)).(((((.....)))))((((((...)))).))))))..... ( -30.10, z-score = -1.09, R) >droSim1.chr2R 4503403 95 - 19596830 GGUCGCUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCGCCCAUU----AAAGGACAACUCCUCAAGUGGGCAUCCAAAGGAUCGCCGCGAUCUC ((((((....(((...((((....))))....)))(((....(((((((----..((((....))))..)))))))((((...))))))).)))))).. ( -37.40, z-score = -3.74, R) >droSec1.super_1 3487274 95 - 14215200 GGUCGCUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCGCCCAUU----AAAGGACAACUCCUCAAGUGGGCAUCCAAAGGAUCGCCGCGAUCUC ((((((....(((...((((....))))....)))(((....(((((((----..((((....))))..)))))))((((...))))))).)))))).. ( -37.40, z-score = -3.74, R) >consensus GGUCGCUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCGCCCAUU____AAAGGACAACUCCUCAAGUGGGCAUCCAAAGGAUCGCCGCGAUCUC ((((((.......(((..((((((.....))))))....................((((....))))...)))(((((((...)))).))))))))).. (-27.04 = -27.16 + 0.12)
| Location | 5,873,207 – 5,873,315 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.40 |
| Shannon entropy | 0.49536 |
| G+C content | 0.53233 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -17.97 |
| Energy contribution | -19.14 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5873207 108 + 21146708 CACUUGAGGAGUUGUCCUUUAAUGGGUGAGGCGCGGAAGUGCAGG-ACUUCCUGGUGUCCUUGAACGACCGGAGCUGAUCUAAAUACGGAUCGGAAUUGGGAUCGAAAU- ((((((((((....)))))....)))))..((((....))))(((-((........)))))....(((((.((.(((((((......)))))))..)).)).)))....- ( -36.30, z-score = -1.82, R) >droAna3.scaffold_13266 4526875 85 - 19884421 ----------------------CACUCGAGUUGCGGAAGUCCUGCCACUUCCUGGU--CCUUGAGAGGGCCUU-CUAAUCUCAAUCAGGAUCUGAAGCAGAUUUCACAAU ----------------------.......((((..((((((.(((((..(((((((--...(((((.......-....))))))))))))..))..))))))))).)))) ( -24.10, z-score = -0.56, R) >droYak2.chr2L 18501198 108 + 22324452 CACUUGAGGAGUGGUCCUUUAACGGGAGACGCGCGGAAGUGCAGG-ACUUCCUGGUGUCCUUGACGGACCGGAGCAGCUCUAGAUCUGGAUCUGAAUCCGGAUCGGAAU- .......((((((((((.(((..((((.((((((....))))(((-....))).)).))))))).)))))......))))).(((((((((....))))))))).....- ( -44.80, z-score = -2.42, R) >droSim1.chr2R 4503429 107 + 19596830 CACUUGAGGAGUUGUCCUUUAAUGGGCGAGGCGCGGAAGUGCAGG-ACUUCCUGGUGUCCUUGAGCGACCGGGGCAGAUCUUAAUAGGGAUCGGAAUCGGGAUUGGAU-- (.((..((((.((((((......)))))).((((....))))(((-....)))....))))..)).).((((..(.((((((....)))))).)..))))........-- ( -40.40, z-score = -1.98, R) >droSec1.super_1 3487300 107 + 14215200 CACUUGAGGAGUUGUCCUUUAAUGGGCGAGGCGCGGAAGUGCAGG-ACUUCCUGGUGUCCUUGAGCGACCGGAGCAGAUCUAAAUAGGGAUCGGAAUCGGGAUUGGAU-- (.((..((((.((((((......)))))).((((....))))(((-....)))....))))..)).).((((..(.(((((......))))).)..))))........-- ( -39.00, z-score = -1.82, R) >consensus CACUUGAGGAGUUGUCCUUUAAUGGGCGAGGCGCGGAAGUGCAGG_ACUUCCUGGUGUCCUUGAGCGACCGGAGCAGAUCUAAAUAGGGAUCGGAAUCGGGAUUGGAA__ .....(((((....))))).......((((((((((((((......))))))..))).)))))..(((((.((...(((((......)))))....)).)).)))..... (-17.97 = -19.14 + 1.17)

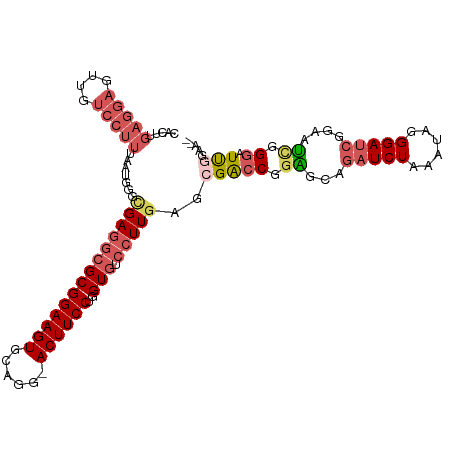
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:27 2011