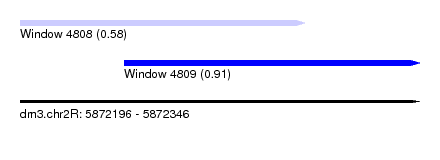
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,872,196 – 5,872,346 |
| Length | 150 |
| Max. P | 0.911574 |

| Location | 5,872,196 – 5,872,303 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Shannon entropy | 0.31529 |
| G+C content | 0.53344 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -20.90 |
| Energy contribution | -22.70 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5872196 107 + 21146708 GAUUCACACGGCUGGCGGGGGGGAUCAACGUUUGAUCAGAAACCGCCGGCGGCUGUUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGGCAGAUUACUGCUGGC---- ..........((((((((....(((((.....))))).....))))))))(((((....)))))......((((.((((..(((.......)))..))))))))...---- ( -42.10, z-score = -4.01, R) >droEre2.scaffold_4929 18144262 97 - 26641161 GAUUCACACGGCUGGCGGG---GAUGCCCGUUUGAUCCAAACCCGCCGCCUG---UUAAUGGCCACAAAAAGCAAGUGACAUGCAAAUUAUACGGAUUAUGGA-------- (((((....(((.((((((---..((...........))..)))))))))((---(((...((........))...)))))............))))).....-------- ( -26.50, z-score = -0.02, R) >droYak2.chr2L 18500199 105 + 22324452 AACUCACCCGGCUGGCGGG---GAUCCCCGUGUGAUCAAAAACCGCCGGCUG---CUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGGCGGAUUACCAGCAAGCGGG ......(((((((((((((---....))))............((((((((((---....))))).......(((.......)))......)))))....)))))...)))) ( -41.40, z-score = -2.43, R) >droSim1.chr2R 4502441 103 + 19596830 GAUUCACACGGCUGGCGGGGGGGAUCACCGUUUGAUCAGAAACCGCCGUCG----UUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGCCAGAUUAAUGGUGGC---- .......(((((.(((((....(((((.....))))).....)))))))))----).....(((.......(((.......)))......((((......)))))))---- ( -31.10, z-score = -0.97, R) >droSec1.super_1 3486315 103 + 14215200 GAUUCACACGGCUGGCGGGGGGGAUCACCGUUUGAUCAGAAACCGCCGUCG----UUAACGGCGACAAAAAGCAAGUGACAUGCAAAUUAGCCAGAUUAAUGGUGGC---- ...((((...((((((((....(((((.....))))).....)))))((((----(.....)))))....)))..))))...........((((......))))...---- ( -33.70, z-score = -1.97, R) >consensus GAUUCACACGGCUGGCGGGGGGGAUCACCGUUUGAUCAGAAACCGCCGGCGG___UUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGCCAGAUUAAUGGUGGC____ ...((((...((((((((....(((((.....))))).....)))))((((........)))).......)))..))))...........((((......))))....... (-20.90 = -22.70 + 1.80)

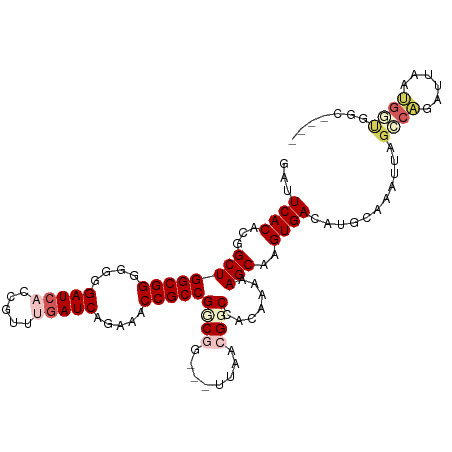
| Location | 5,872,235 – 5,872,346 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.56998 |
| G+C content | 0.51745 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -16.53 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
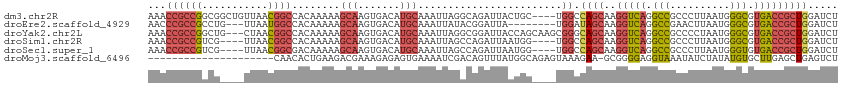
>dm3.chr2R 5872235 111 + 21146708 AAACCGCCGGCGGCUGUUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGGCAGAUUACUGC----UGGCCAGCAAGGUCAGGCCGCCCUUAAUGGGCGUGACCGCUGGAUCU .....(((((((((((....)))))..........(((((..(((.......)))..)))))))----))))((((..(((((...(((((.....))))))))))))))..... ( -48.60, z-score = -3.13, R) >droEre2.scaffold_4929 18144298 104 - 26641161 AACCCGCCGCCUG---UUAAUGGCCACAAAAAGCAAGUGACAUGCAAAUUAUACGGAUUA--------UGGAUAGCAAGGUCAGGCCGAACUUAAUGGGCGUGACCGCUGGAUCU ..((.((.((.((---(((...((........))...))))).))...............--------..........(((((.(((.(......).))).))))))).)).... ( -29.10, z-score = -0.49, R) >droYak2.chr2L 18500235 112 + 22324452 AAACCGCCGGCUG---CUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGGCGGAUUACCAGCAAGCGGGCAGCAAGGUCAGGCCGCCCCUAAUGGGCGUGACCGCUGGAUCU ...((((((((((---....))))).......(((.......)))......)))))....(((((..((.....))..(((((...(((((.....))))))))))))))).... ( -48.40, z-score = -2.80, R) >droSim1.chr2R 4502480 107 + 19596830 AAACCGCCGUCG----UUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGCCAGAUUAAUGG----UGGCCAGCAAGGUCAGGCCGCCCUUAAUGGGCGUGACCGCUGGAUCU ...(((((((.(----(((...((........))...))))..((......)).......))))----)))(((((..(((((...(((((.....))))))))))))))).... ( -39.10, z-score = -2.12, R) >droSec1.super_1 3486354 107 + 14215200 AAACCGCCGUCG----UUAACGGCGACAAAAAGCAAGUGACAUGCAAAUUAGCCAGAUUAAUGG----UGGCCAGCAAGGUCAGGCCGCCCUUAAUGGGUGUGACCGCUGGAUCU ...(((((((((----(.....))))).....(((.......))).................))----)))(((((..(((((...(((((.....))))))))))))))).... ( -39.40, z-score = -2.41, R) >droMoj3.scaffold_6496 4439274 93 + 26866924 ---------------------CAACACUGAAGACGAAAGAGAGUGAAAAUCGACAGUUUAUGGCAGAGUAAAGAA-GCGGGGAGGUAAAUAUCUAUAUGUGCUUGAGCUGAGUCU ---------------------.........((((......((.......))..(((((((.((((.(((......-))..(((........)))...).))))))))))).)))) ( -15.40, z-score = 0.47, R) >consensus AAACCGCCGGCG____UUAACGGCCACAAAAAGCAAGUGACAUGCAAAUUAGACAGAUUAAUGG____UGGCCAGCAAGGUCAGGCCGCCCUUAAUGGGCGUGACCGCUGGAUCU ...((((((...........))))........(((.......)))........................)).((((..(((((.(((..........))).)))))))))..... (-16.53 = -17.50 + 0.97)
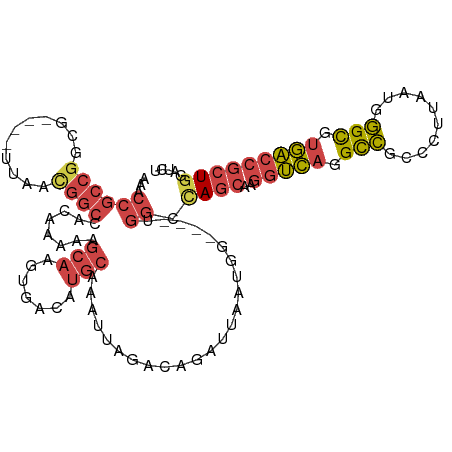
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:26 2011