| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,861,623 – 5,861,756 |
| Length | 133 |
| Max. P | 0.510488 |

| Location | 5,861,623 – 5,861,756 |
|---|---|
| Length | 133 |
| Sequences | 11 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Shannon entropy | 0.48470 |
| G+C content | 0.51296 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5861623 133 + 21146708 GUCCUGUCGUCGUUGUUUU--CCUGUGAUUUGACAUGUCUGUUAGCAGGAUGCCUGACCCUGAGGCCGAGCCC---------UGGUCUCAGU-GU--CCACUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGACUACUG-CUACU (((..((((..(.......--.)..))))..)))........((((((...........(((((((((.....---------))))))))).-((--(((((((......((((((.....)))))).)))))))))..)))-))).. ( -45.10, z-score = -2.00, R) >droSim1.chr2R 4491802 133 + 19596830 GUCCUGUCGUCGUUGUUUU--CCUGUGAUUUGACAUGUCUGUUAGCAGGAUGCCUGACCCUGAGGCCGAGUCC---------UGGACUCAGU-GU--CCACUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGACUACUG-CUACU ((((.(.(.(((......(--(((((.....((....)).....)))))).((((.......)))))))).).---------.)))).((((-((--(((((((......((((((.....)))))).))))))))).))))-..... ( -44.40, z-score = -1.64, R) >droSec1.super_359 7858 133 + 13297 GUCCUGUCGUCGUUGUUUU--CCUGUGAUUUGACAUGUCUGUUAGCAGGAUGCCUGACCCUGAGGCCGAGUCC---------UGGACUCAGU-GU--CCACUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGACUACUG-CUACU ((((.(.(.(((......(--(((((.....((....)).....)))))).((((.......)))))))).).---------.)))).((((-((--(((((((......((((((.....)))))).))))))))).))))-..... ( -44.40, z-score = -1.64, R) >droYak2.chr2L 18488500 133 + 22324452 GUCCUGUCGUCGUUGUUUU--CCCGUGAUUUGACAUGUCUGUUAGCAGGACGCCUGACCCUGAGUUCGAGUCC---------UGAACUCUAU-GG--CCAGUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGACUACUG-CUACU (((((((.((((..(....--.)..)))).(((((....))))))))))))(((.......(((((((.....---------)))))))...-))--)(((((.((((((((((((.....))))))...)))))).)))))-..... ( -42.30, z-score = -1.75, R) >droEre2.scaffold_4929 18133697 135 - 26641161 GUCCUGUCGUCGUUGUUUU--CCCGUGAUUUGACAUGUCUGUUAGCAGGAUGCCUGACCCUGAGGCCGGGUCC---------UGAACUCAGU-GUGUCCAGUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGACUACUG-CUACU (((..((((.((.......--..))))))..)))........((((.((((((..(((((.......)))))(---------((....))).-))))))((((.((((((((((((.....))))))...)))))).)))))-))).. ( -42.40, z-score = -0.76, R) >droAna3.scaffold_13266 4516491 131 - 19884421 GUCCUGUCGUCGAUGGUUU---CUGUGAUUUGACAUGUCUGUUAGCAGGAUGCCCG---CUGACCCUGAGUCC---------UGAGAUCUUA-GGCCUCUGUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGAAU-CUGGCUACC (((..((((..((.....)---)..))))..)))........((((.(((((((((---(((((...(((.((---------(((....)))-)).))).......(((......)))....))))))).)))...))-)).)))).. ( -40.20, z-score = -0.92, R) >dp4.chr3 8870007 129 - 19779522 GUCCUGUCGUCGUUGUUUU--CCCGUGAUUUGACAUGUCUGUUAGCAGGAUGCCGUUGCCUGACCCUGAGCCA---------GAGGACCCUA-------UGUGUUCCACUUUGAUGUGAUUCGUCAAUGCGGUGGACUACUG-CUACC (((..((((.((.......--..))))))..)))........((((((....(((..((.((((...((((((---------.((....)).-------)).))))(((......)))....))))..))..)))....)))-))).. ( -34.90, z-score = -0.59, R) >droPer1.super_4 929771 129 - 7162766 GUCCUGUCGUCGUUGUUUU--CCCGUGAUUUGACAUGUCUGUUAGCAGGAUGCCGUUGCCUGACCCUGAGCUA---------GAGGACCCUA-------CGUGUUCCACUUUGAUGUGAUUCGUCAAUGCGGUGGACUACUG-CUACC (((..((((.((.......--..))))))..)))........((((((....(((..((.((((...((((..---------.((....)).-------...))))(((......)))....))))..))..)))....)))-))).. ( -33.40, z-score = -0.07, R) >droWil1.scaffold_180700 29134 146 + 6630534 GUCCUGUCGUCG-UUGUUUU-CUUAUGAUUUGACAUAUCUGUUGGCAGGACGCUUGACCCUGAACCGAACUACACACAGGGAUGAGCUGAGAUGUUUUGGGUGUUCCACUUUGAUGUGAUUCGUCAAUGUGGUAGACUACUGGCUACC (((((((((.((-.((((.(-(....))...))))....)).)))))))))(((((.(((((..............))))).)))))......(((...(((.(.((((.((((((.....)))))).)))).).)))...))).... ( -45.94, z-score = -1.75, R) >droVir3.scaffold_12875 1332666 113 + 20611582 CCUCUGUCGUCGGUCUGUCAGCCUGUGAUUUGACAUGUUUGUUAGCUAGGC--------CUGACCCUGAGUUG---------------------------GUGUUCCACUUUGAUGUGAUUCGUCAAUGCGGUGGACUACCGGCUACC .........((((...(((((((((.(...(((((....))))).))))).--------))))).))))((((---------------------------(((.((((((((((((.....))))))...)))))).))))))).... ( -39.20, z-score = -2.44, R) >droGri2.scaffold_15245 8664901 117 - 18325388 UUCUUGUCGUCGGCUGUUGU-GCUGUGAUUUGACAUGUUUGUUAGCAGGGCGCUUGACCCUGA------------------------------GUUUUGUGUGUUCCACUUUGAUGUGAUUCGUCAAUGCGGUGGACUACCGGCUACC .((..((((.((((......-))))))))..)).........((((.((((((..(((.....------------------------------)))..))))..((((((((((((.....))))))...))))))...)).)))).. ( -34.60, z-score = -0.89, R) >consensus GUCCUGUCGUCGUUGUUUU__CCUGUGAUUUGACAUGUCUGUUAGCAGGAUGCCUGACCCUGAGCCCGAGUCC_________UGGACUCAGU_GU__CCAGUGUUCCACUUUGAUGUGAUUCGUCAGUGCGGUGGACUACUG_CUACC (((((((.((((.............)))).(((((....)))))))))))).....................................................((((((((((((.....))))))...))))))............ (-20.74 = -20.37 + -0.38)

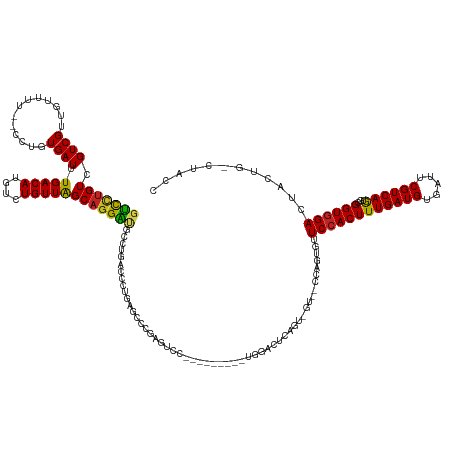

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:23 2011