
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,825,445 – 5,825,536 |
| Length | 91 |
| Max. P | 0.573932 |

| Location | 5,825,445 – 5,825,536 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 64.56 |
| Shannon entropy | 0.69293 |
| G+C content | 0.53064 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -7.91 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
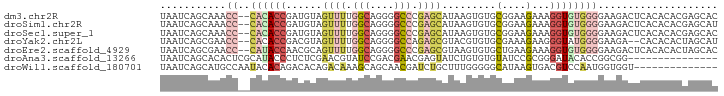
>dm3.chr2R 5825445 91 - 21146708 UAAUCAGCAAACC--CACACCGAUGUAGUUUUGGCAGGGGCCCGAGCAUAAGUGUGCGGAAGAAAGGUGUGGGGAAGACUCACACACGAGCAC ......((...((--(((((..((((...((.(((....))).))))))..))))).)).......(((((((.....)))))))....)).. ( -29.60, z-score = -2.03, R) >droSim1.chr2R 4455731 91 - 19596830 UAAUCAGCAAACC--CACACCGAUGUAGUUUUGGCAGGGGCCCGAGCAUAAGUGUGCGGAAGAAAGGUGUGGGGAAGACUCACACACGAGCAU ......((...((--(((((..((((...((.(((....))).))))))..))))).)).......(((((((.....)))))))....)).. ( -29.60, z-score = -2.08, R) >droSec1.super_1 3435662 91 - 14215200 UAAUCAGCAAACC--CACACCGAUGUAGUUUUGGCAGGGGCCCGAGCAUAAGUGUGCGGAAGAAAGGUGUGGGGAAGACUCACACACGAGCAC ......((...((--(((((..((((...((.(((....))).))))))..))))).)).......(((((((.....)))))))....)).. ( -29.60, z-score = -2.03, R) >droYak2.chr2L 18451357 89 - 22324452 UAAUCAGCGAACC--CACACCGACGUAGUUUUGGCAGGGGCCAGAGCGUACGUGUGCGAAAGAAGGGUAUGGGGAAGA--CACACACUAGCAU ......((...((--((.(((.(((((((((((((....)))))))).)))))...(....)...))).))))..((.--......)).)).. ( -29.30, z-score = -2.68, R) >droEre2.scaffold_4929 18098015 91 + 26641161 UAAUCAGCGAACC--CAUACCAACGCAGUUUUGGCAGGGGCCCGAGCGUAAGUGUGCUGAAGAAAGGUGUGGGGAAGACUCACACACUAGCAC ...((((((..((--(...((((.......))))...))).....((....)).)))))).(..(((((((((.....))))))).))..).. ( -28.50, z-score = -1.19, R) >droAna3.scaffold_13266 15329833 78 - 19884421 UAAUCAGCACACUCGCAUACCCUCUCGAACGUAUCCGACGAACGAGUAUCUGUGUGUAUCCGCGGGAUACACCGGCGG--------------- ..............((((((...((((..(((.....)))..)))).....))))))..((((.((.....)).))))--------------- ( -21.50, z-score = -0.51, R) >droWil1.scaffold_180701 3027007 79 + 3904529 UAAUCAGCAUGCCAAUACACAGACACAGACAAAGCAGCAACGAUCUGCUUUGGGGGCAUAAGUGACGUCCAAUGGUGGU-------------- ..((((.(((...................((((((((.......)))))))).((((.........)))).))).))))-------------- ( -19.00, z-score = -0.49, R) >consensus UAAUCAGCAAACC__CACACCGACGUAGUUUUGGCAGGGGCCCGAGCAUAAGUGUGCGGAAGAAAGGUGUGGGGAAGACUCACACACGAGCAC ...............((((((......((((.(((....))).)))).........(....)...))))))...................... ( -7.91 = -8.67 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:20 2011