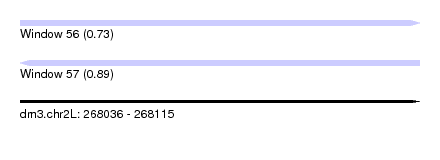
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 268,036 – 268,115 |
| Length | 79 |
| Max. P | 0.894823 |

| Location | 268,036 – 268,115 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 75.53 |
| Shannon entropy | 0.34760 |
| G+C content | 0.48252 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -12.65 |
| Energy contribution | -15.43 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
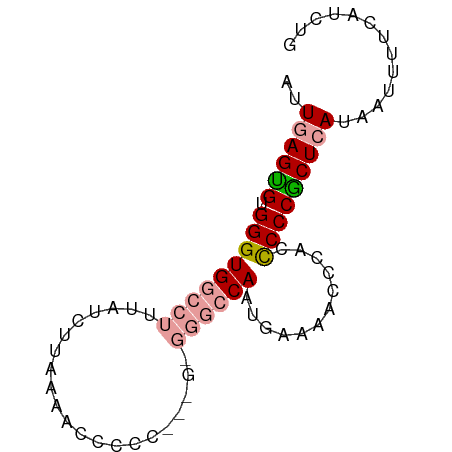
>dm3.chr2L 268036 79 + 23011544 CAUAGGAAAAUUAUUAGCGGGAGAGGAUUUUCAUUGGCCCACCAAGGGGGUUUUAAGAAAAAGGGCACCCACACUCAAU ......................(((..(((((...(((((.(....))))))....))))).((....))...)))... ( -15.50, z-score = 0.15, R) >droEre2.scaffold_4929 319110 78 + 26641161 CAGAUGAAAAUAAUGAGCGGGG-UGGGUUUUCAUUGGCCCGCGCGGGGGAUUUUAAGAUAAAGGCCACCCGCGCUCAAU .............(((((...(-((((((......)))))))(((((((.(((((...))))).)).)))))))))).. ( -32.20, z-score = -3.16, R) >droYak2.chr2L 262041 71 + 22324452 CGGAUGAAAGUUAUGAGUGGGGGUGGGUUUUCAUUGG--------AGGGGCUUUAAGUUAAAGGCCACCCACACUCAAU ..(((....))).((((((.((((((.((((..((((--------((...))))))...)))).)))))).)))))).. ( -26.70, z-score = -3.73, R) >consensus CAGAUGAAAAUUAUGAGCGGGGGUGGGUUUUCAUUGGCCC_C___GGGGGUUUUAAGAUAAAGGCCACCCACACUCAAU .............((((((.((((((.((((..(((((((......)))...))))...)))).)))))).)))))).. (-12.65 = -15.43 + 2.78)



| Location | 268,036 – 268,115 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 75.53 |
| Shannon entropy | 0.34760 |
| G+C content | 0.48252 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -10.83 |
| Energy contribution | -11.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 268036 79 - 23011544 AUUGAGUGUGGGUGCCCUUUUUCUUAAAACCCCCUUGGUGGGCCAAUGAAAAUCCUCUCCCGCUAAUAAUUUUCCUAUG .......(((((.(....(((((......((((....).))).....)))))....).)))))................ ( -15.80, z-score = -0.95, R) >droEre2.scaffold_4929 319110 78 - 26641161 AUUGAGCGCGGGUGGCCUUUAUCUUAAAAUCCCCCGCGCGGGCCAAUGAAAACCCA-CCCCGCUCAUUAUUUUCAUCUG ..((((((.((((((..(((((..........(((....)))...)))))...)))-)))))))))............. ( -24.22, z-score = -2.45, R) >droYak2.chr2L 262041 71 - 22324452 AUUGAGUGUGGGUGGCCUUUAACUUAAAGCCCCU--------CCAAUGAAAACCCACCCCCACUCAUAACUUUCAUCCG ..((((((.((((((.(((((...)))))...(.--------.....).....)))))).))))))............. ( -18.70, z-score = -3.78, R) >consensus AUUGAGUGUGGGUGGCCUUUAUCUUAAAACCCCC___G_GGGCCAAUGAAAACCCACCCCCGCUCAUAAUUUUCAUCUG ..((((((.(((((((((.....................))))))............)))))))))............. (-10.83 = -11.83 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:26 2011