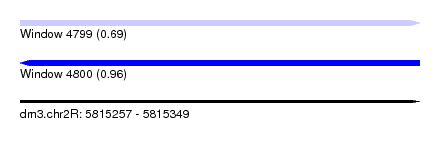
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,815,257 – 5,815,349 |
| Length | 92 |
| Max. P | 0.958647 |

| Location | 5,815,257 – 5,815,349 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.19 |
| Shannon entropy | 0.42527 |
| G+C content | 0.47892 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.19 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5815257 92 + 21146708 -------GCCGAAUGACCGUAAUC-GUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGC-AAAUCU--------GCGGU-GAUUAUCGCCUUC-GAAUGGCUUUGUUUGUU------- -------(((((((.(((((((((-((......))))).(((((((......)))))))-.....)--------)))))-.))).(((....)-)).))))..........------- ( -33.10, z-score = -2.29, R) >droSim1.chr2R 4445697 92 + 19596830 -------GCCGAAUGACCGUUAUC-GUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGC-AAAUCU--------GCGGU-GAUUAUCGCCAUC-GAAUGGCUUUGUUUGCU------- -------(((((((((......))-))))...((((((.(((((((......)))))))-.((((.--------.....-))))))))))...-....)))..........------- ( -32.70, z-score = -1.79, R) >droSec1.super_1 3425646 91 + 14215200 -------GCCGAAUGACCUUUAUC-GUU-AUCGGCGAUCGCCGUGGCUAAGUCCACGGC-AAAUCU--------GCGGU-GAUUAUCGCCAUC-GAAUGGCUUUGUUUGCU------- -------(((((.((((.......-)))-))))))(((((((((((......)))).((-(....)--------)))))-))))...(((((.-..)))))..........------- ( -31.90, z-score = -1.96, R) >droYak2.chr2L 18441130 92 + 22324452 -------GACGAAUAAACGUUAUC-GUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGC-AAAUCU--------GCGGU-GAUUAUCGCCUUG-GAAUAGCUUUGUUUGCU------- -------..((((((((.(((((.-.((((..((((((.(((((((......)))))))-.((((.--------.....-)))))))))).))-)))))))))))))))..------- ( -32.10, z-score = -2.57, R) >droEre2.scaffold_4929 18087699 99 - 26641161 -------GACGAAUAGGCGUUAUC-AUUUAUCGGCGAUCGCCGUGGCUAAGUCCGCGGC-AAAUCU--------GCGCU-GAUUAUCGCCUUC-GAAUGGCUUUGUUUGCUGUUUGCU -------.......(((((.....-....(((((((...(((((((......)))))))-......--------.))))-)))...))))).(-(((((((.......)))))))).. ( -33.60, z-score = -1.28, R) >droAna3.scaffold_13266 15320086 94 + 19884421 -------GCCAAGCUAUCUCAAGCUAUUUUUCGAUAAUCGCAGUGGCCAAGUCCAAAGC-AAAUCU--------AUCCC-GAUUACGGCGAUCUGAAUUGCUUUGUUUGUU------- -------((((((((......))))......((.....))...))))((((..((((((-((.((.--------(((((-(....))).)))..)).))))))))))))..------- ( -24.50, z-score = -2.23, R) >droPer1.super_4 881277 109 - 7162766 ACCGAAAUGUGUGUGUGUGUUAUC-GUUAAUCAAUUAUCGCCGUGGCUAAGUCCAAAACUAAGUCCAAAAUCGAGCGAUAGAUUAUCGCCUUU-CAAUGGCUUUGUUUGUU------- ..(((((..(......)..)).))-).(((((...(((((((((((((.(((.....))).)).)))....)).)))))))))))..(((...-....)))..........------- ( -20.40, z-score = -0.19, R) >consensus _______GCCGAAUGACCGUUAUC_GUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGC_AAAUCU________GCGGU_GAUUAUCGCCUUC_GAAUGGCUUUGUUUGCU_______ ................................(((((.((((((((......)))))))............................(((........)))...).)))))....... (-15.38 = -15.19 + -0.20)
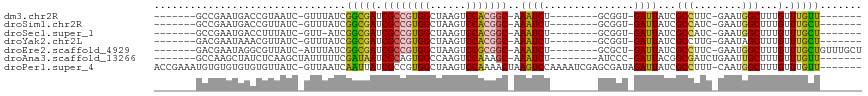
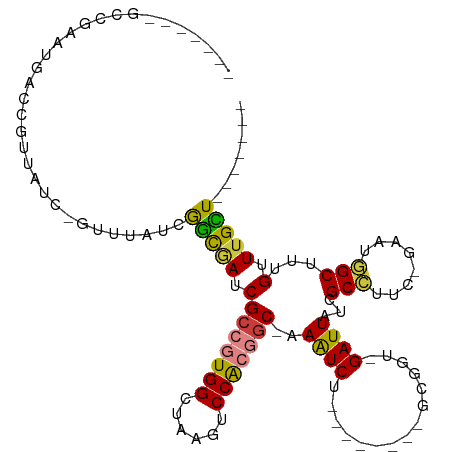
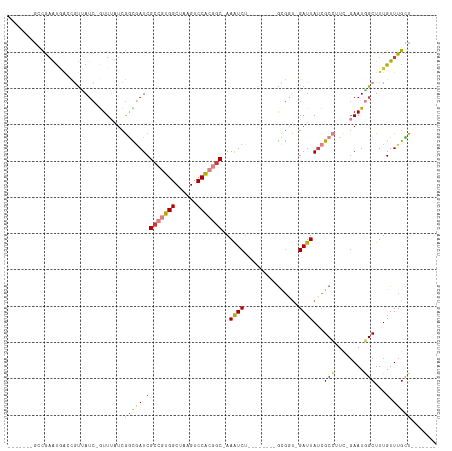
| Location | 5,815,257 – 5,815,349 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.19 |
| Shannon entropy | 0.42527 |
| G+C content | 0.47892 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -14.58 |
| Energy contribution | -15.36 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5815257 92 - 21146708 -------AACAAACAAAGCCAUUC-GAAGGCGAUAAUC-ACCGC--------AGAUUU-GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAAC-GAUUACGGUCAUUCGGC------- -------..........(((....-...((((((((((-.....--------.))))(-(((((((......)))))))))))))).......-(((....)))....)))------- ( -30.40, z-score = -2.82, R) >droSim1.chr2R 4445697 92 - 19596830 -------AGCAAACAAAGCCAUUC-GAUGGCGAUAAUC-ACCGC--------AGAUUU-GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAAC-GAUAACGGUCAUUCGGC------- -------.((.......(((((..-.)))))(.....)-...))--------.((((.-(((((((......)))))))))))(((((....(-(....)).....)))))------- ( -31.60, z-score = -2.61, R) >droSec1.super_1 3425646 91 - 14215200 -------AGCAAACAAAGCCAUUC-GAUGGCGAUAAUC-ACCGC--------AGAUUU-GCCGUGGACUUAGCCACGGCGAUCGCCGAU-AAC-GAUAAAGGUCAUUCGGC------- -------.((.......(((((..-.)))))(.....)-...))--------.((((.-(((((((......)))))))))))(((((.-...-(((....)))..)))))------- ( -31.30, z-score = -2.72, R) >droYak2.chr2L 18441130 92 - 22324452 -------AGCAAACAAAGCUAUUC-CAAGGCGAUAAUC-ACCGC--------AGAUUU-GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAAC-GAUAACGUUUAUUCGUC------- -------(((.......)))....-...((((((((((-.....--------.))))(-(((((((......))))))))))))))(((((((-(....))))))))....------- ( -32.10, z-score = -4.19, R) >droEre2.scaffold_4929 18087699 99 + 26641161 AGCAAACAGCAAACAAAGCCAUUC-GAAGGCGAUAAUC-AGCGC--------AGAUUU-GCCGCGGACUUAGCCACGGCGAUCGCCGAUAAAU-GAUAACGCCUAUUCGUC------- .((.....)).............(-(((((((...(((-(.((.--------.((((.-((((.((......)).))))))))..)).....)-)))..)))))..)))..------- ( -26.60, z-score = -0.96, R) >droAna3.scaffold_13266 15320086 94 - 19884421 -------AACAAACAAAGCAAUUCAGAUCGCCGUAAUC-GGGAU--------AGAUUU-GCUUUGGACUUGGCCACUGCGAUUAUCGAAAAAUAGCUUGAGAUAGCUUGGC------- -------..(((.((((((((.((..(((.(((....)-)))))--------.)).))-))))))...)))((((..((.....((((........))))....)).))))------- ( -27.00, z-score = -2.38, R) >droPer1.super_4 881277 109 + 7162766 -------AACAAACAAAGCCAUUG-AAAGGCGAUAAUCUAUCGCUCGAUUUUGGACUUAGUUUUGGACUUAGCCACGGCGAUAAUUGAUUAAC-GAUAACACACACACACAUUUCGGU -------..........(((....-...)))..(((((((((((.((....(((.((.((((...)))).)))))))))))))...))))).(-((.................))).. ( -20.53, z-score = -1.39, R) >consensus _______AGCAAACAAAGCCAUUC_GAAGGCGAUAAUC_ACCGC________AGAUUU_GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAAC_GAUAACGGUCAUUCGGC_______ ............................((((((((((...............))))..(((((((......))))))).))))))................................ (-14.58 = -15.36 + 0.78)

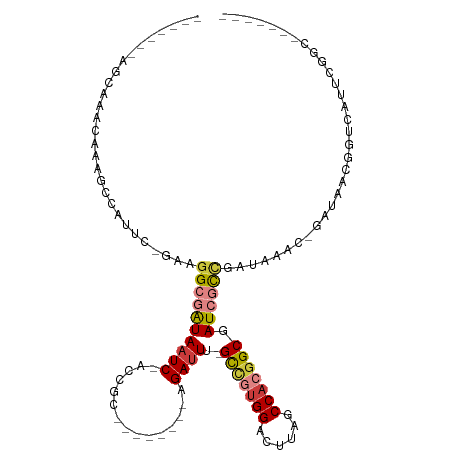
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:18 2011