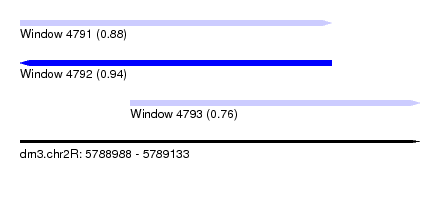
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,788,988 – 5,789,133 |
| Length | 145 |
| Max. P | 0.939156 |

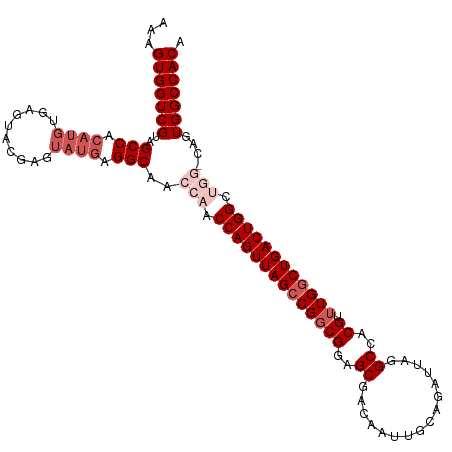
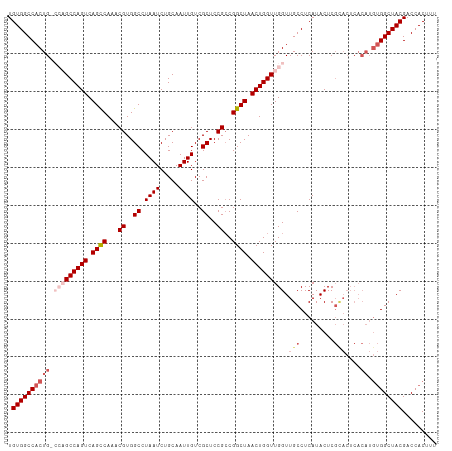
| Location | 5,788,988 – 5,789,101 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Shannon entropy | 0.13577 |
| G+C content | 0.55565 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -36.61 |
| Energy contribution | -39.01 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
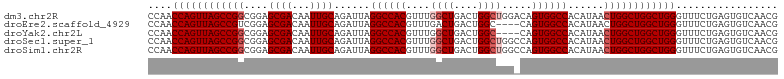
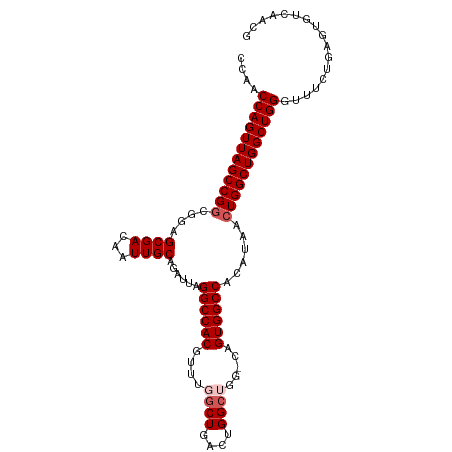
>dm3.chr2R 5788988 113 + 21146708 AAAGUGGUCGUAGCCACAUGUGAGUACGAGUAUGAGGCAACCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGGACAGUGGCCACA ...(((((((..(((.((((..........)))).)))..(((.((((((((((((((..((................))..)).)))))))))))).)))....))))))). ( -45.29, z-score = -2.81, R) >droEre2.scaffold_4929 18061502 97 - 26641161 AAAGUGGUCGUAGCC------------GAGUAUGAGGCAACCAACCAGUUAGCCGUCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGACUGACUGGC----CAGUGGCCACA ...((((((((((..------------........((((((......))).)))((((...))))..))))......(((((.(((.....)))))))----)...)))))). ( -32.30, z-score = -1.35, R) >droYak2.chr2L 18414575 109 + 22324452 AAAGUGGUCGUAGCCACAUGUGAGUACGAGUAUGAGGCAACCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGC----CAGUGGCCACA ...((((((((.(((.((((..........)))).))).))...((((((((((((((..((................))..)).)))))))))))).----....)))))). ( -40.59, z-score = -1.92, R) >droSec1.super_1 3399792 113 + 14215200 AAAGUGGUCGUAGCCACAUGUGAGUGCGAGUAUGAGGCAACCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGGCCAGUGGCCACA ...(((((((..(((.((((..........)))).)))..(((.((((((((((((((..((................))..)).)))))))))))).)))....))))))). ( -44.59, z-score = -1.77, R) >droSim1.chr2R 4419854 113 + 19596830 AAAGUGGUCGUAGCCACAUGUGAGUGCGAGUAUGAGGCAACCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGGCCAGUGGCCACA ...(((((((..(((.((((..........)))).)))..(((.((((((((((((((..((................))..)).)))))))))))).)))....))))))). ( -44.59, z-score = -1.77, R) >consensus AAAGUGGUCGUAGCCACAUGUGAGUACGAGUAUGAGGCAACCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGG_CAGUGGCCACA ...(((((((..(((.((((..........)))).)))..(((.((((((((((((((..((................))..)).)))))))))))).)))....))))))). (-36.61 = -39.01 + 2.40)

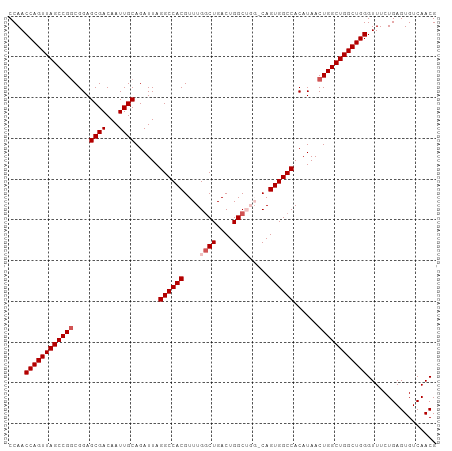
| Location | 5,788,988 – 5,789,101 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Shannon entropy | 0.13577 |
| G+C content | 0.55565 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -32.12 |
| Energy contribution | -34.00 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5788988 113 - 21146708 UGUGGCCACUGUCCAGCCAGUCAGCCAAACGUGGCCUAAUCUGCAAUUGUCGCUCCGCCGGCUAACUGGUUGGUUGCCUCAUACUCGUACUCACAUGUGGCUACGACCACUUU .((((((((((((((((((((.((((...((..((.((((.....))))..))..))..)))).))))))))).(((.........)))...))).))))))))......... ( -40.10, z-score = -2.57, R) >droEre2.scaffold_4929 18061502 97 + 26641161 UGUGGCCACUG----GCCAGUCAGUCAAACGUGGCCUAAUCUGCAAUUGUCGCUCCGACGGCUAACUGGUUGGUUGCCUCAUACUC------------GGCUACGACCACUUU ((.(((.((..----((((((.((((....(..(......)..)....((((...)))))))).))))))..)).))).))...((------------(....)))....... ( -34.00, z-score = -2.13, R) >droYak2.chr2L 18414575 109 - 22324452 UGUGGCCACUG----GCCAGUCAGCCAAACGUGGCCUAAUCUGCAAUUGUCGCUCCGCCGGCUAACUGGUUGGUUGCCUCAUACUCGUACUCACAUGUGGCUACGACCACUUU ((.(((.((..----((((((.((((...((..((.((((.....))))..))..))..)))).))))))..)).))).))...(((((..((....))..)))))....... ( -39.40, z-score = -2.38, R) >droSec1.super_1 3399792 113 - 14215200 UGUGGCCACUGGCCAGCCAGUCAGCCAAACGUGGCCUAAUCUGCAAUUGUCGCUCCGCCGGCUAACUGGUUGGUUGCCUCAUACUCGCACUCACAUGUGGCUACGACCACUUU .((((((((((((((((((((.((((...((..((.((((.....))))..))..))..)))).))))))))))(((.........)))....)).))))))))......... ( -43.60, z-score = -2.87, R) >droSim1.chr2R 4419854 113 - 19596830 UGUGGCCACUGGCCAGCCAGUCAGCCAAACGUGGCCUAAUCUGCAAUUGUCGCUCCGCCGGCUAACUGGUUGGUUGCCUCAUACUCGCACUCACAUGUGGCUACGACCACUUU .((((((((((((((((((((.((((...((..((.((((.....))))..))..))..)))).))))))))))(((.........)))....)).))))))))......... ( -43.60, z-score = -2.87, R) >consensus UGUGGCCACUG_CCAGCCAGUCAGCCAAACGUGGCCUAAUCUGCAAUUGUCGCUCCGCCGGCUAACUGGUUGGUUGCCUCAUACUCGCACUCACAUGUGGCUACGACCACUUU .((((((((...(((((((((.((((...((..((.((((.....))))..))..))..)))).))))))))).(((.........))).......))))))))......... (-32.12 = -34.00 + 1.88)

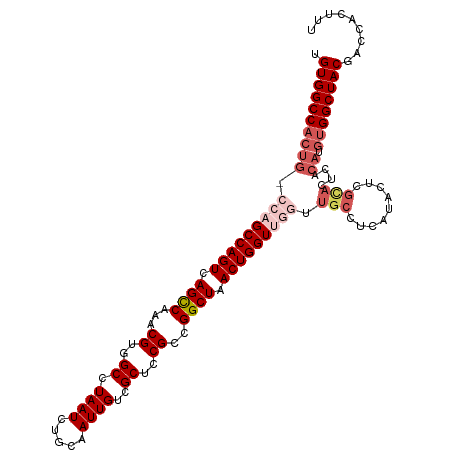
| Location | 5,789,028 – 5,789,133 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.75 |
| Shannon entropy | 0.05599 |
| G+C content | 0.57241 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -37.08 |
| Energy contribution | -37.88 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5789028 105 + 21146708 CCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGGACAGUGGCCACAUAACUGGCUGGCUGGGUUUCUGAGUGUCAACG ....((((((((((((....((((...))))......(((((((((..(((....)))..))).))))))......))))))))))))................. ( -46.10, z-score = -3.17, R) >droEre2.scaffold_4929 18061530 101 - 26641161 CCAACCAGUUAGCCGUCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGACUGACUGGC----CAGUGGCCACAUAACUGGCUGGCUGGGUUUCUGAGUGUCAACG ....(((((((((((((((.((((...))))...((((((...)))))))))))((((----(...))))).......))))))))))................. ( -34.60, z-score = -0.83, R) >droYak2.chr2L 18414615 101 + 22324452 CCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGC----CAGUGGCCACAUAACUGGCUGGCUGGGUUUCUGAGUGUCAACG ....((((((((((((....((((...))))......((((((...(((((....)))----))))))))......))))))))))))................. ( -42.20, z-score = -2.34, R) >droSec1.super_1 3399832 105 + 14215200 CCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGGCCAGUGGCCACAUAACUGGCUGGCUGGGUUUCUGAGUGUCAACG ....((((((((((((....((((...))))......((((((...(((((........)))))))))))......))))))))))))................. ( -42.20, z-score = -1.46, R) >droSim1.chr2R 4419894 105 + 19596830 CCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGGCCAGUGGCCACAUAACUGGCUGGCUGGGUUUCUGAGUGUCAACG ....((((((((((((....((((...))))......((((((...(((((........)))))))))))......))))))))))))................. ( -42.20, z-score = -1.46, R) >consensus CCAACCAGUUAGCCGGCGGAGCGACAAUUGCAGAUUAGGCCACGUUUGGCUGACUGGCUGG_CAGUGGCCACAUAACUGGCUGGCUGGGUUUCUGAGUGUCAACG ....((((((((((((....((((...))))......((((((..(..(((....)))..)...))))))......))))))))))))................. (-37.08 = -37.88 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:12 2011