| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,743,985 – 5,744,083 |
| Length | 98 |
| Max. P | 0.852365 |

| Location | 5,743,985 – 5,744,083 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.78 |
| Shannon entropy | 0.46395 |
| G+C content | 0.35951 |
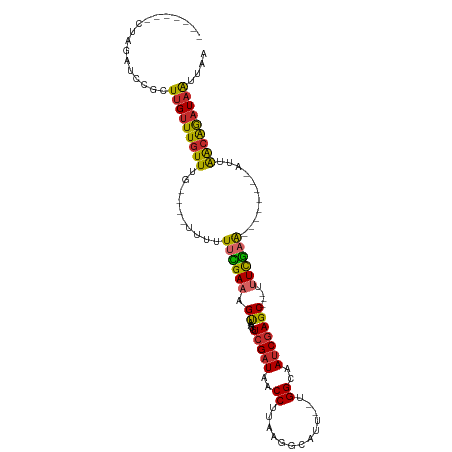
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -12.15 |
| Energy contribution | -11.97 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
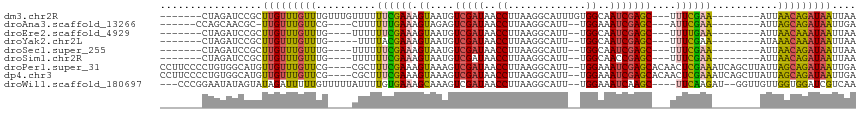
>dm3.chr2R 5743985 98 - 21146708 -------CUAGAUCCGCUUGUUUGUUUGUUUGUUUUUUCGAAAGUAAUGUCGAUAACCUUAAGGCAUUUGUGGCAAUCGAGC---UUUCGAA--------AUUAACAGAUAAUUAA -------..................(((((((((.((((((((((....(((((..((.............))..)))))))---)))))))--------)..))))))))).... ( -23.72, z-score = -2.27, R) >droAna3.scaffold_13266 10865001 92 + 19884421 ------CCAGCAACGC-UUGUUUGUUCG----CUUUUUUGAAAGUAGAGUCGAUAACCUUAAGGCAUU--UGGAAAUCGAGC---AUUCGAA--------AUUAGCAGAUAAUUGA ------...((.....-(((..((((((----..((((..((.((.(((........)))...)).))--..)))).)))))---)..))).--------....)).......... ( -16.80, z-score = 0.55, R) >droEre2.scaffold_4929 18019101 92 + 26641161 -------CUAGAUCCGCUUGUUUGUUUG----UUUUUUCGAAAGUAAUGUCGAUAACCUUAAGGCAUU--UGGCAAUCGAGC---UUUUGAA--------AUUAACAAAUAAUUAA -------..............(((((((----((.((((((((((..((((((...((....))...)--))))).....))---)))))))--------)..))))))))).... ( -22.30, z-score = -2.74, R) >droYak2.chr2L 18370123 91 - 22324452 -------CUAGAUCCGCUUGUUUGUUUG-----UUUUACGAAAGUAAUGUCGAUAACCUUAAGGCAUU--UGGCAAUCGAGC---UUUCGAA--------AUAAACAAAUAAUUAA -------..........(((((((((((-----(((..(((((((..((((((...((....))...)--))))).....))---)))))))--------)))))))))))).... ( -24.60, z-score = -3.63, R) >droSec1.super_255 14594 92 - 18018 -------CUAGAUCCGCUUGUUUGUUUG----UUUUUUCGAAAGUAAUGUCGAUAACCUUAAGGCAUU--UGGCAAUCGAGC---UUUCGAA--------AUUAACAGAUAAUUAA -------..............(((((((----((.((((((((((..((((((...((....))...)--))))).....))---)))))))--------)..))))))))).... ( -24.50, z-score = -3.13, R) >droSim1.chr2R 4381697 92 - 19596830 -------CUAGAUCCGCUUGUUUGUUUG----UUUUUUCGAAAGUAAUGUCGAUAACCUUAAGGCAUU--UGGCAACCGAGC---UUUCGAA--------AUUAACAGAUAAUUAA -------..............(((((((----((.((((((((((..((((((...((....))...)--))))).....))---)))))))--------)..))))))))).... ( -24.50, z-score = -3.18, R) >droPer1.super_31 286129 110 + 935084 CCUUCCCCUGUGGCAUGUUGUUUGUUCG----CGCUUUCGAAAGUAAAGUCGAUAACCUUAAGGCAUU--UGGAAAUCGAGCACAACUCGAAAUCAGCUUAUUAGCAGAUAAUUGA ..((((...(((.(.((..(((...(((----.((((((....).)))))))).)))..)).).))).--.)))).(((((.....))))).(((.(((....))).)))...... ( -25.70, z-score = -0.72, R) >dp4.chr3 17140747 110 - 19779522 CCUUCCCCUGUGGCAUGUUGUUUGUUCG----CGCUUUCGAAAGUAAAGUCGAUAACCUUAAGGCAUU--UGGAAAUCGAGCACAACUCGAAAUCAGCUUAUUAGCAGAUAAUUGA ..((((...(((.(.((..(((...(((----.((((((....).)))))))).)))..)).).))).--.)))).(((((.....))))).(((.(((....))).)))...... ( -25.70, z-score = -0.72, R) >droWil1.scaffold_180697 554115 105 + 4168966 ---CCCGGAAUAUAGUAUAGAUUUUUGUUUUUAUUUUGUGAAAGCAAAGUCGAUAACCUUAAGGCAUU--UGGAAAUCAAGC----UUCAAGAU--GGUUGUUGGUGGAUCGUCAA ---....................(((((((((((...)))))))))))(.((((.(((...((.((((--(.(((.......----))).))))--).))...)))..)))).).. ( -21.50, z-score = -0.47, R) >consensus _______CUAGAUCCGCUUGUUUGUUUG____UUUUUUCGAAAGUAAUGUCGAUAACCUUAAGGCAUU__UGGCAAUCGAGC___UUUCGAA________AUUAACAGAUAAUUAA .................(((((((((..........((((((.......(((((..((.............))..)))))......))))))...........))))))))).... (-12.15 = -11.97 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:07 2011