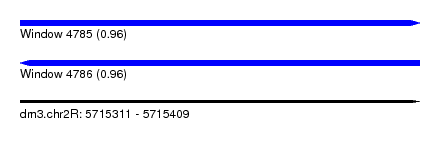
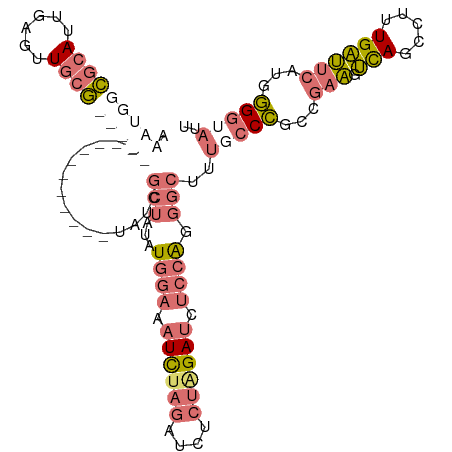
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,715,311 – 5,715,409 |
| Length | 98 |
| Max. P | 0.963071 |

| Location | 5,715,311 – 5,715,409 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.17 |
| Shannon entropy | 0.57396 |
| G+C content | 0.43827 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -12.75 |
| Energy contribution | -14.50 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5715311 98 + 21146708 AAUUCCCCAUGAAUCAAAGGCUGACUUCGGCGGGCAAAGCCCUGGAGAUCUAGAGAUCUAGAUUUCCAUAUAGCGAUA----------------CGCAACUCAAUGCGCCAUUU ..................(((........((((((...))))(((((((((((....)))))))))))....))....----------------.(((......)))))).... ( -31.50, z-score = -2.78, R) >droSim1.chr2R_random 1755133 114 + 2996586 AAUACCCCAUGAAUCAACGGCUGACUUCGGCGGGCAAAGCCCUGGAGAUCCAGAGAUCUAGAUUUCCAUAUAGCGAUAGGAUACGAUACAAUAGCGCAACUCAAUGCGCCAUUU .........((.(((....((((....))))((((...))))((((((((.((....)).))))))))................))).))...(((((......)))))..... ( -31.10, z-score = -1.75, R) >droSec1.super_1 3324210 113 + 14215200 AAUACCCCAUGAAUCAAAGGCUGACUUCGGCGGGCAAAGCC-UGGAGAUCCAGAGAUCUAGAUUUCCAUAUAGCAAUACGAUACGAUACAAUAGCGCAACUCAAUGCGCCAUUU .........((.(((....((((....))))((((...)))-)(((((((.((....)).))))))).................))).))...(((((......)))))..... ( -28.30, z-score = -1.67, R) >droGri2.scaffold_15112 955221 92 + 5172618 ----ACUUUUGUGCCAAAAUGUAGACUUGGCAGAAAAUGGACCUGCU-UCUGUAUGAUUGAAUUUUCAGGAAUGAAU-----------------UUCAAGCUAAUCCAACUUUG ----..((((.((((((.........)))))).))))((((...((.-((.....))(((((..((((....)))).-----------------)))))))...))))...... ( -20.00, z-score = -1.38, R) >consensus AAUACCCCAUGAAUCAAAGGCUGACUUCGGCGGGCAAAGCCCUGGAGAUCCAGAGAUCUAGAUUUCCAUAUAGCAAUA________________CGCAACUCAAUGCGCCAUUU ....(((..(((((((.....))).))))..)))....((..(((((((((((....)))))))))))....))...................(((((......)))))..... (-12.75 = -14.50 + 1.75)

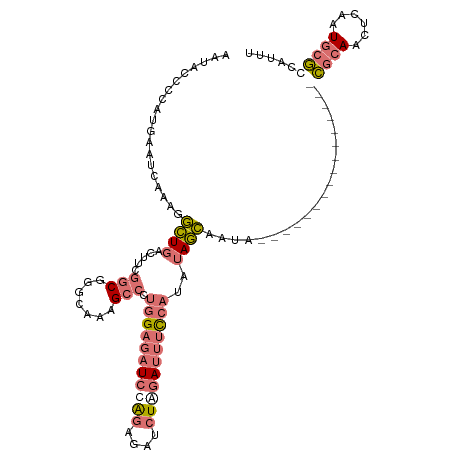
| Location | 5,715,311 – 5,715,409 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 63.17 |
| Shannon entropy | 0.57396 |
| G+C content | 0.43827 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -14.46 |
| Energy contribution | -19.27 |
| Covariance contribution | 4.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 5715311 98 - 21146708 AAAUGGCGCAUUGAGUUGCG----------------UAUCGCUAUAUGGAAAUCUAGAUCUCUAGAUCUCCAGGGCUUUGCCCGCCGAAGUCAGCCUUUGAUUCAUGGGGAAUU .....(((((......))))----------------).((.((((.((((.((((((....)))))).))))((((...))))..(((((.....)))))....)))).))... ( -32.00, z-score = -1.52, R) >droSim1.chr2R_random 1755133 114 - 2996586 AAAUGGCGCAUUGAGUUGCGCUAUUGUAUCGUAUCCUAUCGCUAUAUGGAAAUCUAGAUCUCUGGAUCUCCAGGGCUUUGCCCGCCGAAGUCAGCCGUUGAUUCAUGGGGUAUU .(((((((((......))))))))).....(((((((((.((....((((.((((((....)))))).))))((((...)))))).(((.((((...)))))))))))))))). ( -41.90, z-score = -2.92, R) >droSec1.super_1 3324210 113 - 14215200 AAAUGGCGCAUUGAGUUGCGCUAUUGUAUCGUAUCGUAUUGCUAUAUGGAAAUCUAGAUCUCUGGAUCUCCA-GGCUUUGCCCGCCGAAGUCAGCCUUUGAUUCAUGGGGUAUU .(((((((((......)))))))))((((.(((......))).))))(((.((((((....)))))).))).-(((((((.....))))))).(((((........)))))... ( -36.60, z-score = -1.96, R) >droGri2.scaffold_15112 955221 92 - 5172618 CAAAGUUGGAUUAGCUUGAA-----------------AUUCAUUCCUGAAAAUUCAAUCAUACAGA-AGCAGGUCCAUUUUCUGCCAAGUCUACAUUUUGGCACAAAAGU---- ......(((((..(((((((-----------------.((((....))))..)))).((.....))-)))..)))))((((.(((((((.......))))))).))))..---- ( -20.30, z-score = -1.74, R) >consensus AAAUGGCGCAUUGAGUUGCG________________UAUCGCUAUAUGGAAAUCUAGAUCUCUAGAUCUCCAGGGCUUUGCCCGCCGAAGUCAGCCUUUGAUUCAUGGGGUAUU .(((((((((......)))))))))...............(((...((((.((((((....)))))).)))).)))..(((((...(((.(((.....))))))...))))).. (-14.46 = -19.27 + 4.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:07 2011