
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,688,859 – 5,688,955 |
| Length | 96 |
| Max. P | 0.553815 |

| Location | 5,688,859 – 5,688,955 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.00 |
| Shannon entropy | 0.60012 |
| G+C content | 0.41045 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.44 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553815 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 5688859 96 + 21146708 ------CGGUGCAUGUGUAUGUAUCC-GAAUCUGUAGUUUUUGGCUUGCCAUAUUGUGUGUGUAUCAUAUUUAUGACAGUCUGUCAAUGGCAGAAGCAGGAGC-------- ------((((((((....))))).))-)........(((((((.((((((((...(((((.....)))))...((((.....))))))))))..)))))))))-------- ( -25.20, z-score = -0.43, R) >droMoj3.scaffold_6496 2841136 89 - 26866924 ------------CGUUAUGACUUCUUACCGACUCUGCUUCUUUUUUUUUUUUUUUUUGCUUUAUUUAUGAUUGUCAAUGUCUGCCAAUGACAGGCACGACU---------- ------------.....((((.((...........((....................)).........))..))))..(((((((.......)))).))).---------- ( -11.50, z-score = -0.92, R) >dp4.chr3 14310035 84 + 19779522 ---------------AGUGUGUUUGU-AGGUACAUAGUUUUUGGCCUGCCGUUUUGUGUGUGUUUCAUAUUUAUGGCAGUCUGUCAAUGGCAGCACCAGC----------- ---------------.(.((((((((-....)))..((..((((((((((((...(((((.....)))))..)))))))...)))))..))))))))...----------- ( -24.30, z-score = -1.56, R) >droPer1.super_4 6156617 84 - 7162766 ---------------AGUGUGUUUGU-AGGUACAUAGUUUUUGGCCUGCCGUUUUGUGUGUGUUUCAUAUUUAUGGCAGUCUGUCAAUGGCAGCACCAGC----------- ---------------.(.((((((((-....)))..((..((((((((((((...(((((.....)))))..)))))))...)))))..))))))))...----------- ( -24.30, z-score = -1.56, R) >droAna3.scaffold_13266 12959883 82 + 19884421 ---------------AGUAUAUAUUCUGUUUCUGUAGUUUUCGGCCUGGCAUAUUGUGUGU--UUCAUAUUUAUGACAGUCUGUCAAUGGCAGGAAUGC------------ ---------------............((((((((.(((..(((.(((.((((.((((...--..))))..)))).))).)))..))).))))))))..------------ ( -20.80, z-score = -1.80, R) >droEre2.scaffold_4929 17964101 102 - 26641161 ------CAGUGCCUCUGUAUGUAUUC-GAAUCUGUAGUUUUUGGCUUGCCAUAUUGUGUGU--AUCAUAUUUAUGACAGUCUGUCAAUGGCAGGAGCAGCAGGAGCAGGAG ------...(((..((((.(((..((-(((.........)))))((((((((..((((...--..))))....((((.....)))))))))))).)))))))..))).... ( -27.60, z-score = -0.20, R) >droYak2.chr2L 18313739 100 + 22324452 AAGUGCAUGUGUACGAGUAUGUAUAC-GAAUCUGUAGUUUUUGGCUUGCCAUAUUGUGUGU--AUCAUAUUUAUGACAGUCUGUCAAUGGCAGGAGCAGGAGC-------- ...(((((((((((......))))))-)....))))(((((((.((((((((..((((...--..))))....((((.....))))))))))))..)))))))-------- ( -28.50, z-score = -1.17, R) >droSec1.super_1 3297845 94 + 14215200 ------CAGUGCAUGUUUAUGUAUCU-GAAUCUGUAGUUUUUGGCUUGCCAUAUUGUGUGU--AUCAUAUUUAUGACAGUCUGUCAAUGGCAGGAGCAGGAGC-------- ------((((((((....))))).))-)........(((((((.((((((((..((((...--..))))....((((.....))))))))))))..)))))))-------- ( -26.80, z-score = -1.25, R) >droSim1.chr2R 4326508 94 + 19596830 ------CAGUGCAUGUGUAUGUAUCU-GAAUCUGUAGUUUUUGGCUUGUCAUAUUGUGUGU--AUCAUAUUUAUGACAGUCUGUCAAUGGCAGGAGCAGGAGC-------- ------..((((((....))))))((-(..(((((.....(((((((((((((.((((...--..))))..))))))))...)))))..)))))..)))....-------- ( -25.20, z-score = -0.74, R) >consensus ________GUG_AUGUGUAUGUAUCU_GAAUCUGUAGUUUUUGGCUUGCCAUAUUGUGUGU__AUCAUAUUUAUGACAGUCUGUCAAUGGCAGGAGCAGCAG_________ .............................................(((((((...(((((.....)))))...((((.....))))))))))).................. (-10.70 = -10.44 + -0.26)

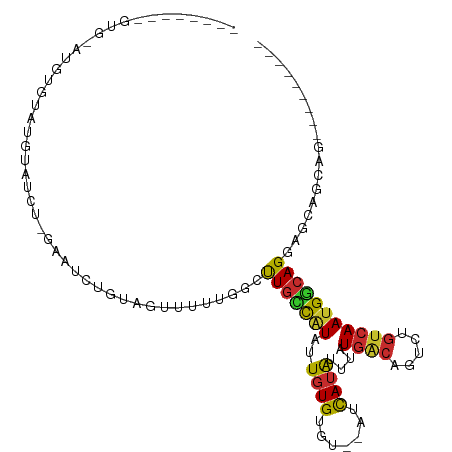

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:11:03 2011