| Sequence ID | dm3.chr2R |
|---|---|
| Location | 5,656,648 – 5,656,764 |
| Length | 116 |
| Max. P | 0.763620 |

| Location | 5,656,648 – 5,656,764 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.69545 |
| G+C content | 0.60510 |
| Mean single sequence MFE | -38.99 |
| Consensus MFE | -15.31 |
| Energy contribution | -16.40 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
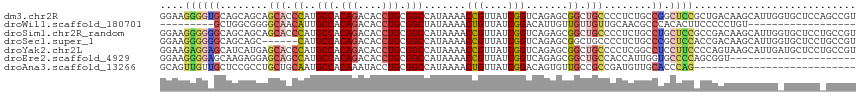
>dm3.chr2R 5656648 116 + 21146708 GGAAGGGGUGCAGCAGCAGCACCCAUGCCACAGACACCUGCGGCCAUAAAACCGUUAUCGGUCAGAGCGGCUGCCCCUCUGCCGGCUCCGCUGACAAGCAUUGGUGCUCCAGCCGU ((.(((((.(((((.((.........(((.(((....))).)))......((((....))))....)).))))).))))).))(((...(((....)))..(((....)))))).. ( -46.00, z-score = -0.51, R) >droWil1.scaffold_180701 3629786 89 + 3904529 ---------GCUGGCGGGGCAACAUUGCCACAGACACCUGCGGCUAUAAAACUGUUAUCGGACAUUGUUGUUGUUGCAACGCCCACACUUCCCCCUGU------------------ ---------...((((..((((((..(((.(((....))).))).((((...((((....))))...))))))))))..))))...............------------------ ( -27.70, z-score = -0.99, R) >droSim1.chr2R_random 1739244 116 + 2996586 GGAAGGGGGGCAGCAGCAGCACCCAUGCCACAGACACCUGCGGCCAUAAAACCGUUAUCGGUCAGAGCGGCUGCCCCUCUGCCUGCUCCGCCGACAAGCAUUGGUGCUCCUGCCGU ((.(((((((((((.((.........(((.(((....))).)))......((((....))))....)).))))))))))).)).((..((((((......)))))).....))... ( -47.80, z-score = -0.72, R) >droSec1.super_1 3265714 110 + 14215200 GGAAGGGGGGCAGCAGC------CAUGCCACAGACACCUGCGGCCAUAAAACCGUUAUCGGUCAGAGCGGCUGCCCCUCUGCCCGCUCCACCGACAAGCAUUGGUGCUCCUGCCGU ((.(((((((((((.((------(..(((.(((....))).)))......((((....))))..).)).)))))))))))..))((..((((((......)))))).....))... ( -48.20, z-score = -1.90, R) >droYak2.chr2L 18281101 116 + 22324452 GGAAGAGGAGCAUCAUGAGCACCCAUGCCACAGACACCUGCGGCCAUAAAACCGUUAUCGGUCAGAGCGGCUGCCCCUCGGCCUCCUUCCCCAGUAAGCAUUGAUGCUCCUGCCGU ((...((((((((((((.((......(((.(((....))).)))......((((....))))....))(((((.....)))))...............)).)))))))))).)).. ( -40.70, z-score = -1.98, R) >droEre2.scaffold_4929 17931097 95 - 26641161 GGAAGGGGAGCAAGAGGAGCAGCCAUGCCACAGACACCUGCGGCCAUAAAACCGUUAUCGGUCAGAGCGGCUGCCACCAUUGGUGCCCCAGCGGU--------------------- ....(((((.(((..(..((((((..(((.(((....))).)))......((((....))))......))))))..)..))).).))))......--------------------- ( -37.80, z-score = -1.60, R) >droAna3.scaffold_13266 12927202 89 + 19884421 GCAGUUGUUGCUCCGCCUGCUGCAAUGCCACAAAUACCUGCGGCCAUAAAACUGUUAUCGGACAGUGUUGCCGCCGAUGUUGCACCCAG--------------------------- (((((.((......))..)))))..(((.(((.....(.(((((......((((((....))))))...))))).).))).))).....--------------------------- ( -24.70, z-score = -0.29, R) >consensus GGAAGGGGAGCAGCAGCAGCACCCAUGCCACAGACACCUGCGGCCAUAAAACCGUUAUCGGUCAGAGCGGCUGCCCCUCUGCCUGCUCCACCGACAAGCAUUG_UGCUCC_GCCGU ((.(((((.((...............(((.(((....))).))).......(((....)))...........)).))))).))................................. (-15.31 = -16.40 + 1.09)

| Location | 5,656,648 – 5,656,764 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.69545 |
| G+C content | 0.60510 |
| Mean single sequence MFE | -44.71 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
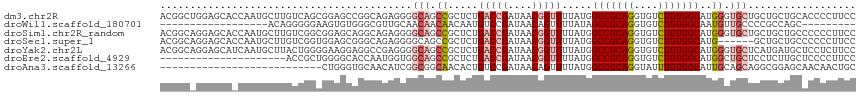
>dm3.chr2R 5656648 116 - 21146708 ACGGCUGGAGCACCAAUGCUUGUCAGCGGAGCCGGCAGAGGGGCAGCCGCUCUGACCGAUAACGGUUUUAUGGCCGCAGGUGUCUGUGGCAUGGGUGCUGCUGCUGCACCCCUUCC .((((.(.((((((.(((((.....(((((((((((((((.((...)).))))).)).....(((((....)))))..))).)))))))))).)))))).).)))).......... ( -50.10, z-score = -0.20, R) >droWil1.scaffold_180701 3629786 89 - 3904529 ------------------ACAGGGGGAAGUGUGGGCGUUGCAACAACAACAAUGUCCGAUAACAGUUUUAUAGCCGCAGGUGUCUGUGGCAAUGUUGCCCCGCCAGC--------- ------------------...(((((..((.(((((((((.........)))))))))...)).((..(((.(((((((....))))))).)))..))))).))...--------- ( -33.60, z-score = -1.73, R) >droSim1.chr2R_random 1739244 116 - 2996586 ACGGCAGGAGCACCAAUGCUUGUCGGCGGAGCAGGCAGAGGGGCAGCCGCUCUGACCGAUAACGGUUUUAUGGCCGCAGGUGUCUGUGGCAUGGGUGCUGCUGCUGCCCCCCUUCC .((((((.((((((..((.(((((((((((((.(((.........))))))))).)))))))))........(((((((....)))))))...)))))).)))))).......... ( -58.50, z-score = -2.10, R) >droSec1.super_1 3265714 110 - 14215200 ACGGCAGGAGCACCAAUGCUUGUCGGUGGAGCGGGCAGAGGGGCAGCCGCUCUGACCGAUAACGGUUUUAUGGCCGCAGGUGUCUGUGGCAUG------GCUGCUGCCCCCCUUCC ..(((((.((((((.....(((((((((((((((((......))..))))))).)))))))).)))......(((((((....)))))))...------))).)))))........ ( -51.50, z-score = -1.29, R) >droYak2.chr2L 18281101 116 - 22324452 ACGGCAGGAGCAUCAAUGCUUACUGGGGAAGGAGGCCGAGGGGCAGCCGCUCUGACCGAUAACGGUUUUAUGGCCGCAGGUGUCUGUGGCAUGGGUGCUCAUGAUGCUCCUCUUCC ..((.((((((((((.......((.((........)).))(((((.(((....(((((....))))).....(((((((....))))))).))).))))).))))))))))))... ( -50.40, z-score = -1.96, R) >droEre2.scaffold_4929 17931097 95 + 26641161 ---------------------ACCGCUGGGGCACCAAUGGUGGCAGCCGCUCUGACCGAUAACGGUUUUAUGGCCGCAGGUGUCUGUGGCAUGGCUGCUCCUCUUGCUCCCCUUCC ---------------------......((((...(((.((..(((((((....(((((....))))).....(((((((....))))))).)))))))..)).)))..)))).... ( -41.80, z-score = -2.31, R) >droAna3.scaffold_13266 12927202 89 - 19884421 ---------------------------CUGGGUGCAACAUCGGCGGCAACACUGUCCGAUAACAGUUUUAUGGCCGCAGGUAUUUGUGGCAUUGCAGCAGGCGGAGCAACAACUGC ---------------------------.....(((((.((((((((.....))).)))))............(((((((....))))))).)))))((((..(......)..)))) ( -27.10, z-score = 0.14, R) >consensus ACGGC_GGAGCA_CAAUGCUUGUCGGCGGAGCAGGCAGAGGGGCAGCCGCUCUGACCGAUAACGGUUUUAUGGCCGCAGGUGUCUGUGGCAUGGGUGCUGCUGCUGCACCCCUUCC ..........................................(((.((.....(((((....))))).....(((((((....)))))))..)).))).................. (-16.40 = -17.30 + 0.90)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:10:59 2011