| Sequence ID | dm3.chr2L |
|---|---|
| Location | 2,436,714 – 2,436,806 |
| Length | 92 |
| Max. P | 0.916483 |

| Location | 2,436,714 – 2,436,806 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 64.94 |
| Shannon entropy | 0.66289 |
| G+C content | 0.52739 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -10.18 |
| Energy contribution | -9.51 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
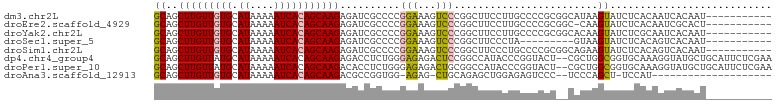
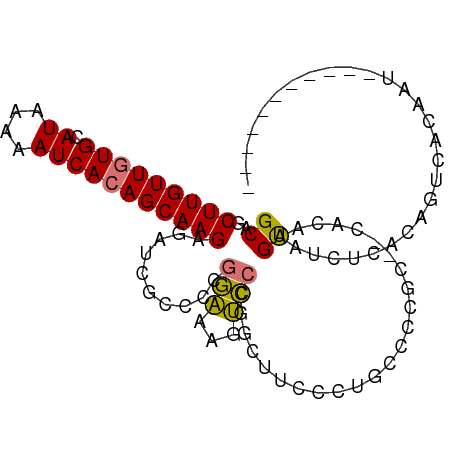
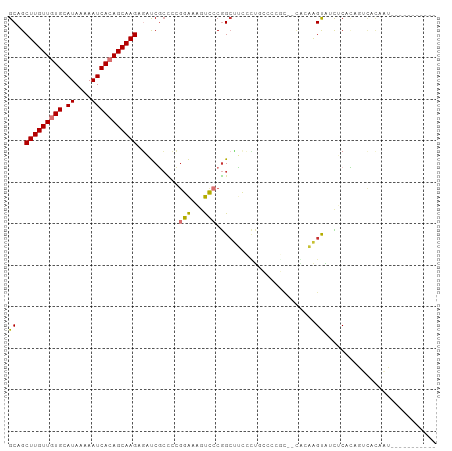
>dm3.chr2L 2436714 92 + 23011544 GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGAGAUCGCCCCGGAAAGUCCCGGCUUCCUUGCCCCGCGGCAUAAGUAUCUCACAAUCACAAU----------- .....((((((((.((....))))))))))(((((.((.((((......))))......((((....))))...)))))))...........----------- ( -23.70, z-score = -1.71, R) >droEre2.scaffold_4929 2480578 91 + 26641161 GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGAGAUCGCCCCGGAAAGUCCCGGCUUCCUUGCCCCGCGGC-CAAGUAUCUCACAAUCGCACU----------- ((...((((((((.((....))))))))))(((((((((.(((........(((......)))))).)))-...).)))))......))...----------- ( -25.80, z-score = -1.83, R) >droYak2.chr2L 2426289 92 + 22324452 GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGAGAUCGCCCCGGAAAGUCCCGGCUUCCUUGCCCCGCGGCACAAGUAUCUCGCAAUCACAAU----------- ((...((((((((.((....))))))))))(((((.((.((((......))))......((((....))))...))))))))).........----------- ( -26.10, z-score = -1.95, R) >droSec1.super_5 610299 83 + 5866729 GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGAGAUCGCCCCGGAAAGUCCCGGCUUCCCUA---------GUAAGUAUCUCACAGUCACAAU----------- .....((((((((.((....))))))))))((((((.....((.((((....)))).))..---------....).)))))...........----------- ( -17.80, z-score = -1.59, R) >droSim1.chr2L 2395646 92 + 22036055 GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGAGAUCGCCCCGGAAAGUCCCGGCUUCCCUGCCCCGCGGCAGAAGUAUCUCACAGUCACAAU----------- .....((((((((.((....))))))))))(((((.((.((((......)))).....(((((....)))))..)))))))...........----------- ( -26.90, z-score = -2.30, R) >dp4.chr4_group4 1701055 101 + 6586962 GCAGCUUGUUAUGCAUAAAAAUCACAGCAAGAGACCUCUGGGAGAGACUCCGGCCAUACCCGGUACU--CGCUGGCGGUGCAAAGGUAUGCUGCAUUCUCGAA (((((......(((((........((((......((....)).(((...((((......))))..))--)))))...))))).......)))))......... ( -30.02, z-score = -0.26, R) >droPer1.super_10 704511 101 + 3432795 GCAGCUUGUUAUGCAUAAAAAUCACAGCAAGACACCUCUGGGAGAGACUGCGGCCAUACCCGGUACU--CGCUGGCGGUGCAAAGGUAUGCUGCAUUCUCGAA .(((..((((.(((............))).))))...)))...((((.((((((.(((((..((((.--((....))))))...))))))))))).))))... ( -35.80, z-score = -1.91, R) >droAna3.scaffold_12913 173103 78 - 441482 GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGACGCCGGUGG-AGAG-CUGCAGAGCUGGAGAGUCCC--UCCCAGCU-UCCAU-------------------- (((((((((((((.((....))))))))....((....)).-.)))-)))).(((((((.(((...)--))))))))-)....-------------------- ( -29.40, z-score = -2.29, R) >consensus GCAGCUUGUUGUGCAUAAAAAUCACAGCAAGAGAUCGCCCCGGAAAGUCCCGGCUUCCCUGCCCCGC__CACAAGUAUCUCACAGUCACAAU___________ ((..(((((((((.((....)))))))))))..........(((...)))........................))........................... (-10.18 = -9.51 + -0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:11:16 2011